Abstract
The interferon-stimulated gene 56 (ISG56) family is induced strongly in response to virus infection, interferons (IFNs) and double-stranded RNA (dsRNA). In the mouse, this family comprises three members, ISG56, ISG54, and ISG49, which are clustered on chromosome 19 and encode the corresponding proteins p56, p54, and p49. Here, we report differential properties of these proteins and their distinct induction patterns in different cell types. All three murine proteins bound to the c-subunit of the translation initiation factor eIF3, but unlike the other members, p49 did not inhibit protein synthesis. Using a newly raised antibody, we demonstrated that both in vitro and in vivo, p49 expression was strongly induced by IFN, dsRNA, and Sendai virus. However, in kidney mesangial cells, as opposed to podocytes, encephalomyocarditis virus, vesicular stomatitis virus, or extracellular dsRNA did not induce any of the p56 family proteins, although they were robustly expressed after Sendai virus infection or dsRNA transfection. Furthermore, protein-specific differences in the regulation of p56 family members became evident in various leukocyte types: all three proteins were induced by IFN in T cells, but in B cells p56 and ISG56 mRNA could not be detected. Similarly, p56 was selectively uninducible in plasmacytoid dendritic cells, whereas in myeloid dendritic cells, all three family members were expressed. These results revealed novel cell type-, inducer-, and gene-specific regulation of the ISG56 family of genes.
The innate immune response to virus infections largely depends on the action of type I interferons (IFN), a group of cytokines mainly consisting of IFN-β and more than 10 subtypes of IFN-α, which induce expression of a large number of genes referred to as IFN-stimulated genes (ISGs) (6, 31, 38), whose respective protein products mediate numerous cellular functions, including antiviral effects (14, 33, 35, 36). The induction of ISGs is predominantly triggered by the IFN-mediated activation of the JaK-Stat pathway. Binding of type I IFN to the IFN-α/β receptor leads to the activation of the receptor-associated kinases, Jak1 and Tyk2, which in turn phosphorylate STAT1 and STAT2. The heterodimer of STAT1/2 then binds to IFN regulatory factor 9 (IRF-9), forming the transcription factor complex ISGF3 (ISG factor 3), which in the nucleus binds to the IFN-stimulated response element (ISRE) in the promoters of ISGs, inducing their transcription (31, 38).
The triggering of the type I IFN system by viruses depends on the detection of viral molecular patterns such as double-stranded RNA (dsRNA) by cellular receptors, resulting in the induction of IFN-α/β genes (20, 34). Examples of such receptors, detecting different forms of RNA as an intermediate or by-product of viral replication, are the endosomal transmembrane protein Toll-like receptor 3 (TLR3) (1) and the cytoplasmic RNA helicases RIG-I and MDA-5 (3, 11, 44, 45). Binding of dsRNA to TLR3 or RIG-I/MDA-5 triggers mutual signaling pathways, leading to the activation of several transcription factors such as IRF-3 and IRF-7 (17), whose activation requires phosphorylation by their kinases TBK1 or IKKɛ, followed by dimerization and nuclear translocation (9, 27, 37). In the nucleus, IRF-3/7 engage transcription of type I IFNs via binding to promoter elements highly similar to the ISREs of IFN-stimulated genes. Not surprisingly, a large number of ISGs have been found to be inducible not only via IFN-activated ISGF3 but also directly by virus-activated IRF-3 (2, 8, 12). This group of genes is referred to as viral stress-inducible genes (VSIGs) (36).
Among the most strongly induced VSIGs (by either IFN or virus infections) is the ISG56 family of genes, comprising four human members (ISG54/IFIT2, ISG56/IFIT1, ISG58/IFIT5, and ISG60/IFIT3) and three members in mice (ISG49/Ifit3, ISG54/Ifit2, and ISG56/Ifit1), clustered on chromosomes 10 and 19, respectively (4, 7, 23, 25, 36, 43). All encoded proteins, designated p54, p56, etc., contain arrays of multiple tetratricopeptide repeats, which are degenerate helix-turn-helix modules mediating protein-protein interactions, often with larger protein complexes (5, 36). Human and mouse p54 and p56 interact with different subunits of translation initiation factor 3 (eIF3), achieving an inhibition of translation. However, since eIF3 is composed of 13 subunits and has multiple functions in translation initiation (15, 16), the mechanisms of inhibition are diverse. Human p54 and p56 bind eIF3e, thereby inhibiting the stabilization of the eIF2/GTP/Met-tRNA ternary complex (13, 18, 40). Mouse p54 and p56, and in addition, human p54, bind to eIF3c and block the formation of the 48S preinitiation complex consisting of the 40S ribosome subunit, the ternary complex, eIF4F, and mRNA (19, 40, 41). Antiviral functions of p56 proteins, exemplified by studies with hepatitis C virus (39), Sindbis virus (46), and human papillomavirus (F. Terenzi, P. Saikia, and G. C. Sen, unpublished results) have only begun to emerge.
We recently reported an unexpected complexity of murine p54 versus p56 protein expression in vivo, depending on stimulus (IFN, dsRNA, or virus infection), tissue type and cell type (42). For example, whereas tail vein injection of dsRNA induced p54 and p56 in the liver and spleen, injection of VSV induced p54 in the spleen but not in the liver, a phenomenon observed at both protein and mRNA levels. Furthermore, the relative induction of p54 versus p56 in organs such as the heart or lungs varies depending on the inducing stimulus. To date, nothing is known about murine p49 protein expression and function due to a lack of reagents in the past. In order to expand our knowledge about p49 and the other murine family members, we first raised an antibody against p49 and examined the expression and inducibility of the protein in vitro and in vivo. In addition, we found p49 not to be a translation inhibitor, although it is capable of interacting with eIF3c. Furthermore, we discovered cell type- and gene-specific differences in the inducibility of p56 family proteins, revealing that murine p56 is not inducible by IFN in B cells and plasmacytoid dendritic cells (pDCs).
MATERIALS AND METHODS
Cell culture.
HT1080 human fibrosarcoma cells (32), RAW 264.7 murine macrophages, wild-type murine embryonic fibroblasts (MEFs), Stat1−/− MEFs (G. Stark, Lerner Research Institute, Cleveland, OH), IRF3−/− MEFs (T. Taniguchi, University of Tokyo, Tokyo, Japan) were all maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Atlanta Biologicals), 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
Mesangial cell and podocyte culture.
Primary mouse mesangial cells were cultured from isolated glomeruli as described previously (22). Conditionally immortalized murine podocytes, which were isolated from mice that express the temperature-sensitive simian virus 40 T antigen (Immortomouse), were as previously described (29).
B- and T-cell isolation.
Single cell suspensions were prepared from murine peripheral blood, which was obtained by cardiac puncture followed by the lysis of red blood cells with Tris ammonium chloride. Then, 106 cells per sample were stimulated with 1,000 U of IFN-β/ml for 24 h and stained for flow cytometry with antibodies to B220, CD3, and p56 family proteins. B cells were isolated from single cell splenocyte suspensions by negative depletion using MACS beads (Miltenyi Biotech) according to the manufacturer's instructions. Briefly, this involved coating non-B cells with biotinylated antibodies (Miltenyi Biotech), followed by binding of these cells to streptavidin-coated beads (Miltenyi Biotech). These bead-covered cells were removed from the culture with a magnet, while B cells flowed through and were collected. Non-B cells (predominantly T cells) adhered to magnetic beads during the B-cell isolation protocol. Following the retention of beads in the B-cell purification column, the column was removed from the magnet, and the bead-bound non-B cells were eluted. Although the purity of the B cells was generally >97%, non-B-cell preparations were less pure, averaging 73% CD3+ cells, both determined by labeling with CD3 and B220 antibodies (BD Pharmingen). Afterward, cells were treated with 1,000 U of IFN-β (Calbiochem)/ml for 24 h.
Dendritic cell culture.
Dendritic cells were generated from precursors in murine bone marrow by culture for 10 days in the presence of 100 ng of Flt3-ligand (Peprotech)/ml to generate cultures containing a mix of myeloid dendritic cells (mDCs) and pDCs, or they were cultured for 10 days in the presence of 20 ng of granulocyte-macrophage colony-stimulating factor (Peprotech)/ml to produce a culture containing predominantly mDCs. After stimulation with 1,000 U of IFN-β/ml for 16 h, flow cytometry was used to distinguish pDCs (CD11clo B220+ CD11b−) and mDCs (CD11chi B220− CD11b+) in conjunction with intracellular flow cytometry (described below) (10, 26).
Flow cytometry.
To stain cells with cell surface markers for flow cytometry, cells were incubated with an antibody to block Fcγ receptors (clone 2.4G2; BD Pharmingen) before the addition of antibodies to CD4, CD8, B220, CD3, CD11b, or CD11c (clones RM4-5, 53-6.7, RA3-6B2, M1/70, 145-2C11, or HL3, respectively, from BD Pharmingen). For intracellular staining, surface-labeled cells were fixed in 1% paraformaldehyde, followed by permeabilization with saponin. Cells were incubated with antibodies to p49, p54, or p56, which were detected with goat anti-rabbit immunoglobulin G labeled with Alexafluor 488 (Molecular Probes). Flow cytometry was conducted on a FACScan flow cytometer (Becton Dickinson), and the data were analyzed by using FlowJo software (TreeStar, Inc.).
Treatments and virus infections.
Recombinant murine IFN-α or IFN-β (Calbiochem) was used at 500 or 1,000 U/ml for the times indicated. Synthetic dsRNA [poly(I)-poly(C) = poly(IC); GE Amersham] was added to the media at 100 μg/ml or cells were transfected at 1 μg/ml using FuGENE 6 (Roche) for 8 h. Virus infections were usually performed at a multiplicity of infection of 10 in DMEM containing 2% fetal bovine serum for 1 h, followed by two washes with phosphate-buffered saline (PBS) and incubation with DMEM plus 10% fetal bovine serum for 8 h. The viruses were encephalomyocarditis virus (EMCV; a gift from R. Silverman, The Lerner Research Institute, Cleveland, OH), vesicular stomatitis virus (VSV; Indiana strain; a gift from A. Banerjee, The Lerner Research Institute, Cleveland, OH), and Sendai virus (SeV; Cantell strain, Charles River, SPAFAS).
Plasmids.
The full-length cDNA of murine ISG49/Ifit3 was obtained by reverse transcription-PCR (RT-PCR) using RNA extracted from IFN-β-treated RAW 264.7 cells and was cloned into Myc-pcDNA3, expressing N-terminally Myc-tagged p49. The ISG49 cDNA was subcloned into pET-15b by PCR for bacterial expression. Expression vectors for murine p56 (41), murine and human Myc-p54 (40, 41), eIF3c (41), and eIF3e (13) were described before. The cDNA of human ISG60/IFIT3 was cloned into Myc-pcDNA3 by RT-PCR of RNA from IFN-treated HT1080 cells. All plasmid transfections were mediated by FuGENE 6 (Roche) according to the manufacturer's instructions.
Protein purification.
Briefly, Escherichia coli BL21(DE3)/pLysS (Novagen) were transformed with pET-15b/ISG49 or ISG56, and expression of His-tagged p49 or p56 was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 6 h at 30°C. Protein purification was done as described previously (41) using Ni-NTA Superflow beads (Qiagen).
Antibodies.
Polyclonal antibody to murine p49 was raised at the Hybridoma Core, The Lerner Research Institute (Cleveland, OH), by injecting bacterially expressed, purified full-length p49 into rabbits. Antibodies to murine p54 and p56 were raised similarly (42). Anti-c-Myc monoclonal antibody 9E10, anti-Flag clone M2 beads, and antibodies to actin/β-actin were from Sigma.
Mice.
Experiments were performed with FVB mice (tissue screenings and kidney immunohistochemistry) and C57BL/6 mice (B and T cells, fluorescence-activated cell sorting analysis) obtained from Taconic Farms. BALB/c mice (for primary mesangial cell preparation) were from The Jackson Laboratory. All mice were used at 8 to 12 weeks of age. Where indicated, mice were injected intravenously (i.v.) 8 h before sampling with 2 × 105 U of recombinant murine IFN-α or 100 μg of dsRNA [poly(IC)], each in 100 μl of phosphate-buffered saline (PBS) or with PBS alone. For kidney immunohistochemistry, mice were injected i.v. with 107 PFU of SeV 8 h before sampling or the same volume of PBS. For mouse organ tissue extraction, mice were sacrificed by CO2 inhalation, and the organs were removed and immediately frozen in liquid nitrogen. Protein extracts from the organs were made as previously described (28), and relative expression levels of p49 and p56 were quantified by using ImageQuant software (Molecular Dynamics).
Immunoprecipitation and immunoblotting.
For immunoprecipitation of p49, 50 μg of whole-cell extract of Myc-p49- or Myc-p56-expressing MEFs were mixed with 2 μl of anti-p49 antibody in 500 μl of radioimmunoprecipitation assay buffer and subjected to immunoprecipitation (40). Immunoprecipitation of Flag-tagged protein has been described before (40). Cell lysis and immunoblotting were done as described previously (24), using lysis buffer with Complete protease inhibitor cocktail (Roche).
Immunofluorescence.
MEFs were grown in four-well chamber slides (LabTek II; Nunc) and treated with 1,000 U of murine IFN-β (Calbiochem)/ml for 16 h. Cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 15 min, and blocked with 10% fetal bovine serum in PBS for 60 min. p49 was labeled using 1:2,000 dilutions of primary anti-p49 and secondary goat anti-rabbit antibodies (Gibco-BRL) labeled with fluorescein isothiocyanate in 1% fetal bovine serum, and cells were examined by using inverted fluorescence microscopy (Leica DM IRB).
Immunohistochemistry.
Kidneys were collected 8 h after i.v. injection with SeV, fixed in 10% formaldehyde, and embedded in paraffin. For antigen retrieval, 4-μm-thick deparaffinized sections were digested in 0.0025% trypsin plus 0.1% CaCl2 in 0.05 M Tris (pH 7.6) at 37°C for 30 min. After blocking with 1% bovine serum albumin in PBS for 60 min, the sections were incubated with anti-p54 antibody (1:2,000) at 4°C overnight. For detection and visualization, the Envision+ kit (Dako) was used, followed by nuclear staining by Mayer's hematoxylin solution (Sigma) and mounting with Cytoseal 60 mounting medium (Richard-Allan Scientific).
In vitro translation inhibition assay.
Using the rabbit reticulocyte lysate system (Promega), 0.5 μg of luciferase mRNA (Promega) was translated in the presence or absence of purified p49 or p56 proteins in 25-μl reactions containing reticulocyte lysate, amino acids, RNase inhibitor, and [35S]cysteine. After 2 h at 30°C, newly synthesized luciferase protein was resolved by subjecting a 5-μl reaction to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and quantifying the radioactive luciferase signals with a Storm 820 phosphorimager (Molecular Dynamics) using ImageQuant software.
RT-PCR.
B cells were stimulated for 4 h with 1,000 U of murine recombinant IFN-β (Calbiochem)/ml, followed by RNA isolation using TRIzol (Invitrogen) according to the manufacturer's instructions; DNA was removed by using DNAfree (Ambion). RT of RNA was performed with a Superscript III kit (Invitrogen); PCRs were driven by Hot Start Taq polymerase (Denville). The RT-PCR primers used (annealing temperature, 52°C) were as follows: ISG49, 5′-GCCGTTACAGGGAAATACTGG and 3′-CCTCAACATCGGGGCTCT; ISG54, 5′-GGGAAAGCAGAGGAAATCAA and 3′-TGAAAGTTGCCATACAGAAG; ISG56, 5′-CAGAAGCACACATTGAAGAA and 3′-TGTAAGTAGCCAGAGGAAGG; and 18S rRNA, 5′-ATTGACGGAAGGGCACCACCAG and 3′-CAAATCGCTCCACCAACTAAGAACG.
RESULTS
p49 binds to eIF3c but does not inhibit translation.
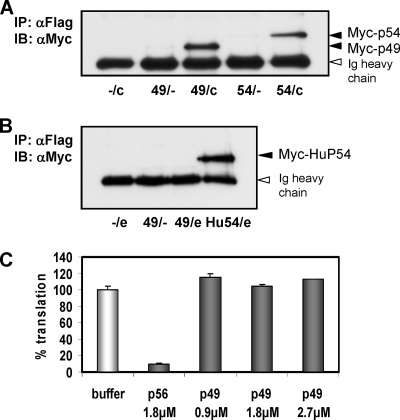
We previously reported murine p54 and p56 as being inhibitors of protein translation initiation, an effect mediated via direct binding to eIF3c (19, 41). We extended our studies by examining these properties of p49. HT1080 cells were cotransfected with expression vectors for Myc-p49 or Myc-p54 as a positive control and Flag-eIF3c (Fig. 1A). Both p49 and p54 were coprecipitated with eIF3c in a Flag immunoprecipitation, demonstrating that like its murine relatives, p49 was capable of binding to eIF3c. Next, interaction between p49 and eIF3e was tested by a similar approach, with human P54 as a positive control (Fig. 1B). In this experiment, p49 did not precipitate with eIF3e, again sharing the binding preferences with its murine relatives. To test the functional consequence of the observed binding of p49 to eIF3c, we used an in vitro translation inhibition assay (40), translating luciferase mRNA in reticulocyte lysate in the presence of various amounts of purified p49 protein (Fig. 1C). Whereas suitable amounts of purified p56 strongly inhibited translation, increasing amounts of p49 had no inhibitory effect at all. These results demonstrated that p49 can bind to eIF3 but does not inhibit its functions in translation initiation in vitro.
FIG. 1.
p49 binds to eIF3c but does not inhibit translation. (A) HT1080 cells were cotransfected with expression vectors for Myc-tagged murine p49 or murine p54 and Flag-tagged eIF3c. After Flag-immunoprecipitation, coprecipitated Myc-tagged proteins were detected by immunoblotting. (B) HT1080 cells were cotransfected with expression vectors for Myc-tagged murine p49 or human p54 and Flag-tagged eIF3e. After Flag immunoprecipitation, coprecipitated Myc-tagged proteins were detected by immunoblotting. (C) Translation inhibition by p49 and p56 was quantified in vitro by using reticulocyte lysate-driven luciferase mRNA translation in the presence of [35S]cysteine. Densitometric averages of polyacrylamide gel electrophoresis-resolved luciferase signal are shown.
Specificity of a newly raised p49 antibody.
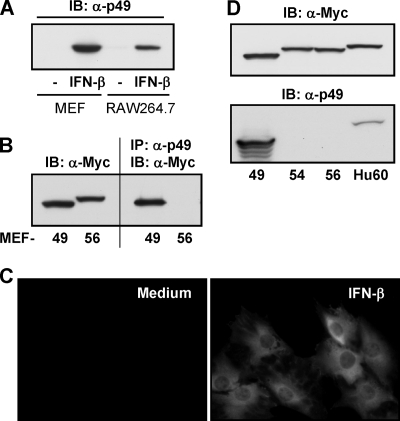
In order to allow for further p49 protein-level experiments in cell culture and in vivo, we raised an antibody by injection of bacterially expressed, purified full-length p49 into rabbits. The specificity of the antiserum was initially tested by immunoblotting of lysates from IFN-β-treated MEFs or RAW 264.7 macrophages (Fig. 2A), yielding a single clean band of the expected molecular mass of 47 kDa in IFN-β-treated lysates, with minor or no constitutive expression of p49. Next, the suitability of the p49 antibody for immunoprecipitation was tested with lysates of MEFs stably expressing Myc-p49 or Myc-p56 (Fig. 2B). The p49 antibody selectively precipitated p49, but not p56. Further confirmation of specificity against other p56 family members was obtained by expressing Myc-tagged p49, p54, p56, and human P60 in HT1080 cells and comparing immunoblots labeled to detect Myc or p49 (Fig. 2D). In the murine family, the p49 antibody recognized exclusively p49, and among the human proteins it recognized, to a minor extent, human P60, which represents the human homolog of p49. These results established the newly raised p49 antibody as a very specific and versatile reagent. Its first use was to determine the subcellular localization of IFN-induced p49 in MEFs (Fig. 2D). Treatment with IFN-β for 18 h resulted in a clearly detectable expression of p49, displaying a diffuse cytoplasmic distribution.
FIG. 2.
Specificity of p49 antibody and cellular localization of p49. (A) The newly raised p49 antibody was used to detect endogenous p49 by immunoblotting after 18 h treatment of MEFs or RAW 264.7 cells with 1,000 U of IFN-β/ml. (B) Immunoprecipitation of p49 was done with cell lysates of MEFs stably expressing Myc-p49 or Myc-p56. Precipitated protein was detected by Myc-immunoblotting. (C) Specificity of p49 antibody was demonstrated with cell lysates from HT1080 cells transfected with vectors for expression of Myc-p49, -p54, or -p56 or Myc-human P60, the homolog of murine p49. (D) Cellular localization of endogenous p49, induced in MEFs by 16 h of treatment with IFN-β, was examined by immunofluorescence.
Induction of p49 by SeV infection requires IFN.
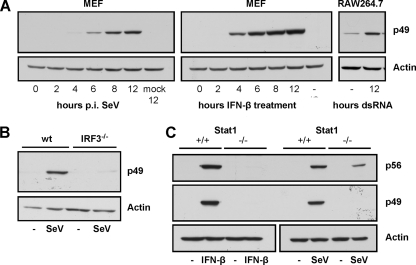
In order to further characterize inducibility of p49 expression by various stimuli, MEFs were infected with SeV (representing a RIG-I activator) or treated with IFN-β. The macrophage line RAW 264.7 was treated with 100 μg of floating dsRNA (representing a TLR3 activator)/ml (Fig. 3A). Both SeV infection and IFN-β treatment induced expression of substantial amounts of p49 starting at 6 h postinfection or after 4 h of treatment, respectively. In RAW 264.7 cells, which displayed a weak constitutive expression of p49, dsRNA treatment led to an elevation of p49 expression by severalfold. Since ISG49 expression is driven by an ISRE in its promoter, we sought to find out whether induction in case of SeV infection could be directly mediated by RIG-I-activated IRF-3, qualifying ISG49 as a VSIG (36). First, wild-type and IRF3−/− MEFs were infected with SeV, and p49 expression was examined (Fig. 3B). In IRF3−/− cells, p49 expression was completely abolished, as expected. Next, wild-type and Stat1−/− MEFs were infected with SeV or treated with IFN-β as a control (Fig. 3C). Whereas IFN-β did not induce expression of either p49 or p56 in Stat1−/− cells, p56 was still inducible by SeV, but surprisingly, p49 was not, demonstrating the requirement for IFN signaling (via the JaK-Stat pathway) for the induction of p49 expression and implying differences in the regulation of p49 and p56 expression.
FIG. 3.
Induction of p49 by SeV infection requires IFN. (A) p49 expression was detected by immunoblotting of cell lysates of either MEFs, after infection with 80 hemagglutinating units (HAU) of SeV/ml or treatment with 1,000 U of IFN-β/ml for the indicated times, or of RAW 264.7 cells, after the addition of dsRNA [100 μg of poly(IC)/ml] to the medium for 12 h. (B) IRF-3 dependence of virus-induced p49 expression was demonstrated by infecting wild-type or IRF3−/− MEFs with 80 HAU of SeV/ml for 16 h and immunoblotting cell lysates, detecting protein expression with p49 or actin antibodies. (C) IFN dependence of virus-induced p49 but not p56 expression was demonstrated by treating wild-type or Stat1−/− MEFs with IFN-β or infecting the cells with 80 HAU of SeV/ml for 16 h and immunoblotting the cell lysates.
Induction of p49 in vivo.
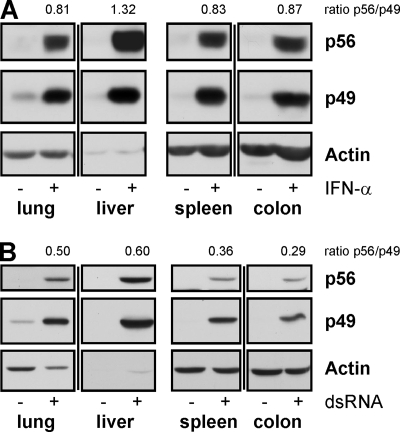
In order to determine whether p49 and p56 were coordinately induced in mouse organs, mice were injected with IFN-α- or dsRNA; 8 h later, the animals were sacrificed, and tissue extracts were examined for p49 and p56 expression (Fig. 4). Organs included the lungs, heart, kidney, liver, spleen, small intestine, colon, brain, and muscles (Fig. 4 and data not shown). In all tissues or organs tested, both stimuli resulted in coordinated expression of both p49 and p56; however, inducer-dependent differences were observed in p56 levels relative to p49. For example, in the spleens and colons of dsRNA-injected mice, the ratios of p56 expression over p49 expression (0.36 and 0.29) were much lower compared to the same ratios (0.83 and 0.87) in IFN-injected mice. On the other hand, in other tissues, such as the lung and liver, these differences were not so prominent (Fig. 4).
FIG. 4.
Induction of p49 and p56 in mice injected with IFN or dsRNA. At 8 h after i.v. injection of IFN-α (A) or dsRNA (B) or PBS, mice were sacrificed and organs were homogenized. Organ extracts (40 μg in panel A and 20 μg in panel B) were examined for expression of p49, p56, or actin by immunoblotting.
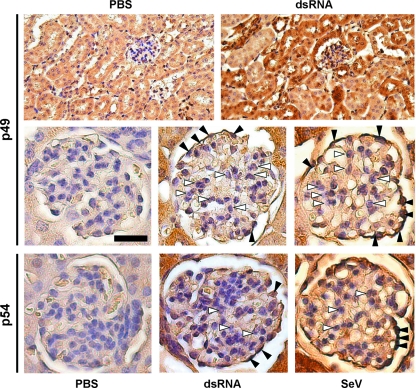
The global inducibility of expression of p56 family members in the mouse prompted us to take a closer look at different cell types rather than whole organs by examining the kidneys of dsRNA- or SeV-injected mice for p49 expression using immunohistochemistry (Fig. 5). The key structures in the renal cortex are the spherical glomeruli, which serve as blood filters. Apart from capillaries, the glomeruli consist of two adjacent cell types, the supporting mesangial cells and the podocytes, which form the plasma filter. The glomeruli are surrounded by tubuli, which convey the plasma filtrate toward the kidney core. Whereas the majority of renal tubular cells, the predominant cell type in the renal cortex, were positive for p49 (Fig. 5, upper panel), within the glomeruli only podocytes were positive for p49 expression after tail vein injection of dsRNA or SeV (Fig. 5, black arrowheads, middle panel); the mesangial cells (Fig. 5, white arrowheads) and capillary cells did not express p49. When kidney sections were stained with p54 or p56 antibody, again only podocytes stained positive for expression, whereas mesangial were negative for both p54 and p56 (Fig. 5, lower panel and data not shown). Similar positive staining for all three proteins in podocytes and tubular cells, but not mesangial cells, was observed after injection of IFN into mice as well (data not shown).
FIG. 5.
Induction of p49 and p54 in the glomeruli of murine kidneys. At 8 h after i.v. injection of dsRNA, SeV, or PBS, mice were sacrificed, kidneys were collected, fixed, and processed for immunohistochemistry to detect p49 (upper and middle panel) and p54 (lower panel). White arrowheads indicate mesangial cells, black arrowheads indicate podocytes in the blood-filtrating glomeruli, located in the renal cortex. Magnifications are ×200 (upper panel) and ×400 (middle and lower panels). Scale bar, 25 μm.
Selective inducibility of p56 family members in mesangial cells.
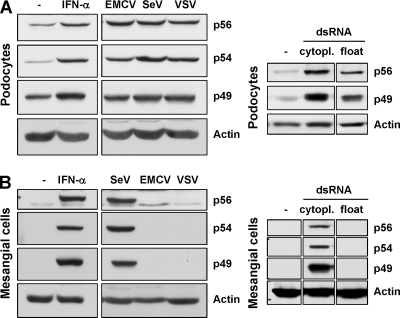
In our in vivo experiments (Fig. 5 and data not shown), mesangial cells did not display expression of p49, p54, or p56 after any of the stimuli used. Therefore, we examined both mesangial cells and podocytes more closely in vitro. Murine podocytes were treated with IFN-α, infected with different viruses (EMCV, SeV, or VSV), or transfected or treated with dsRNA (Fig. 6A). Cell lysates were prepared, and the expression of p49, p54, and p56 was assessed by immunoblotting. In podocytes, all stimuli led to an upregulation of all three proteins, adding to a low constitutive expression level. However, when mesangial cells were treated likewise, expression of the three p56 family proteins was not induced by infection with EMCV or VSV or by treatment with floating dsRNA (Fig. 6B). However, they were robustly induced by IFN-α treatment or SeV infection.
FIG. 6.
Selective inducibility of p56 family members in mesangial cells. Differentiated podocytes (A) and mesangial cells (B) were either treated with 500 U of IFN-α/ml or infected at a multiplicity of infection of 10 with EMCV, SeV, or VSV for 8 h. Expression of p49, p54, and p56 was detected by immunoblotting. Alternatively, cells were exposed to dsRNA [poly(IC)] by either FuGENE 6-mediated transfection (cytopl.) or by adding poly(IC) to the medium (float). In mesangial cells, infection by VSV was verified by detection of P-protein and EMCV infection by observation of development of the cytopathic effect (not shown).
Lack of induction of p56 by IFN in B cells in vitro.
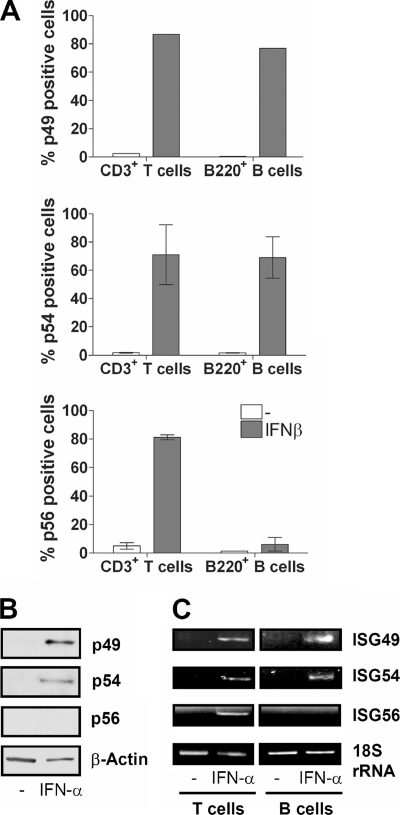
We previously reported that in vivo, B cells do not express p56 after i.v. IFN injection, whereas p54 is induced (42). We wanted to confirm this finding in vitro and examine the inducibility of p49. Peripheral blood cells were isolated from mice; treated with IFN-β for 24 h; and stained with p49, p54, or p56 antibody for flow cytometry, distinguishing B cells (B220+) and T cells (CD3+) (Fig. 7A). Whereas all three proteins were inducibly expressed in ∼75% of CD3+ cells, p56 was not detected in B220+ B cells. Confirmation for this finding was obtained by immunoblotting of the B-cell extracts with antibodies for all three p56 family members (Fig. 7B) and, indeed, both p49 and p54 were present, but p56 was absent in IFN-β-treated B cells. The underlying regulatory mechanism was at the transcriptional level, since the mRNAs for ISG49 and ISG54, but not ISG56, were present in IFN-treated B cells (Fig. 7C).
FIG. 7.
Lack of induction of p56 by IFN in B cells in vitro. (A) Peripheral blood cells were isolated from mice and stimulated with 1,000 U of IFN-β/ml (106 cells/sample). After stimulation (24 h), cells were harvested and stained for flow cytometry. (B) Murine B cells isolated from spleens were untreated or treated with IFN-α for 24 h, and extracts were immunoblotted for the indicated proteins. (C) After 4 h of stimulation of isolated B or T cells with 1,000 U of IFN-α/ml, RNA was isolated. cDNA produced from this RNA provided the template for PCR using primers for ISG49, ISG54, ISG56, and 18S rRNA.
p56 is induced in mDCs but not pDCs.
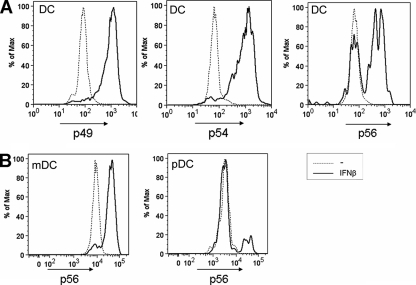
To extend our observation of differential behavior of B cells and T cells to other cells of the immune system, we tested by flow cytometry for p49, p54, and p56 expression in IFN-β-stimulated in vitro-differentiated dendritic cells. We noticed that only a subpopulation of dendritic cells expressed p56, whereas p49 and p54 were expressed in virtually all cells (Fig. 8A). Since the dendritic cells used in these experiments represented a mixture of mDCs and pDCs, we next designed an experiment in which these subtypes were distinguishable. The murine bone-marrow derived precursor cells were in vitro differentiated in the presence of Flt3 ligand, treated with IFN-β, and stained for dendritic cell surface markers in addition to staining for intracellular p56 (Fig. 8B). In virtually all mDCs, p56 expression was detectable, but pDCs were incapable of expressing p56 after treatment with IFN-β, demonstrating profound differences in the regulation of p56 expression in pDCs from that of the other family members.
FIG. 8.
p56 is induced in mDCs but not pDCs. (A) Dendritic cells were differentiated from mouse bone marrow using granulocyte-macrophage colony-stimulating factor. Cells were stimulated for 16 h with 1,000 U of IFN-β/ml before analysis by flow cytometry using antibodies to p49, p54, and p56. (B) Dendritic cells differentiated from bone marrow using Flt3-ligand were stimulated for 16 h with 1,000 U of IFN-β/ml before analysis by flow cytometry. pDC and mDC populations were distinguished by using surface markers, and the p56 expression was detected.
DISCUSSION
We have been studying the inducibility and the functions of the human and murine genes of the ISG56 family (13, 18, 19, 40-42). Historically, these genes have been used by many laboratories as sentinel ISGs for analyzing signaling pathways activated by IFNs, dsRNA, and virus infection. Because these genes are clustered and homologous in sequence, the general assumption has been that they are always coordinately induced and hence the induction properties ascribed to one of these genes are freely applicable to others as well. The results presented here have demonstrated that this simplistic assumption is wrong and that there is profound inducer- and cell type-specific differential regulation of different members of this gene family. Both biosynthetic and functional analyses of the p56 family of proteins have been greatly facilitated by the production of recombinant proteins and raising antibodies against them. We have previously reported the properties of two murine members, p56 and p54 (41, 42); the present study of the third member, p49, completes the basic characterization of all members of the murine ISG56 family.
Antibodies to these murine proteins have been used to monitor their induction in vivo in response to various inducers. They are induced widely in mice infected with various viruses or injected with IFN or dsRNA (42). Functionally, both human and mouse p56 and p54 inhibit translation initiation by binding to different subunits of the translation initiation factor eIF3 (13, 18, 19, 40, 41). Their inhibitory effect on viral protein synthesis has major roles in the antiviral effects of IFN against hepatitis C virus (39; M. Gale, unpublished data) and West Nile virus (Gale, unpublished; M. Diamond, unpublished data) among others. We show here that, in contrast to p56 and p54, p49 does not inhibit translation, although, like the others, it can bind to eIF3c. Further investigation will be required to provide the molecular basis for the observed differential consequences on translation initiation, upon binding of these proteins to eIF3c. Mapping of the respective binding domains may reveal that the differential effect is due to their binding to different regions of eIF3c. This is not an unlikely scenario, considering that p56 and p54 bind to two different regions of eIF3c (41). For identifying cellular functions of p49, we may have to search for additional cellular or viral proteins with which it interacts. Such a search for human p56 has led us to the discovery of its interaction with the human papillomavirus (HPV) E1 protein, which results in a block of replication of the viral DNA (Terenzi et al., unpublished results). As noted by us, the binding specificities of the members of the p56 family are high; for example, HPV E1 does not bind to the other members of the p56 family. Thus, it is reasonable to expect that in the future, we will observe member-specific antiviral effects of these proteins.
The murine ISG56-related genes are clustered on chromosome 19 in a locus of about 100 kb. One copy of ISG54 gene is followed by two alleles of ISG49 gene, a pseudogene of ISG56 and the ISG56 gene. The 5′ promoter-proximal region of all of these genes contains two ISRE elements which respond to the transcriptional induction signals from IFNs, dsRNA, and virus infection. Their physical proximity and the similarity of their promoters led to the expectation that their transcriptional inductions are coordinately regulated. Although this expectation was largely fulfilled by the results presented here, there were notable exceptions, indicating further complexity in the regulation of expression of these genes. The first example of differential regulation is their induction pattern upon SeV infection of MEFs (Fig. 3). As expected, IRF-3 was needed for the induction of p49 (and p56 and p54 [data not shown]), but p49, in contrast to p56, depended on Stat1 as well. The simplest interpretation of our data is that IRF-3 is required for IFN induction, which in turn induces both p56 and p49. However, as reported before, p56 could also be induced by IRF-3 directly in Stat1−/− MEFs, which are unresponsive to IFN (41). Most probably, the induced level of p56 in wild-type MEFs is a composite of the product of direct induction and the product of indirect induction by IFN produced by the infected cells; consequently, the level of p56 in infected Stat1−/− cells was lower than that in wild-type cells. More importantly, our data suggest that SeV-activated IRF-3 cannot induce p49 (Fig. 3C). Further investigation will be needed to examine whether IRF-3 activated by other viruses or dsRNA also fails to induce p49. If this is indeed the case, it will indicate that the ISG49 promoter can respond to type I IFN signaling, which uses ISGF3 as the relevant transcription factor, but not to other signaling pathways, such as RIG-I or TLR3, which use IRF-3 as the transcription factor. Appropriate analyses in the future will reveal whether this property is imparted by the ISREs of ISG49 itself, compared to the ISREs of ISG56. It has been noted before that minor sequence differences in the ISRE can render it unrecognizable by IRF-3, but not ISGF3 (2, 30).
The second notable difference was in the ratios of p56 and p49 induced in specific organs of mice injected with dsRNA. For example, in the colon and in the spleen, the relative induction of p49 was stronger compared to p56 (Fig. 4B). In contrast, in the same tissues, IFN-α induced both proteins with almost equal ratios (Fig. 4A). These observations point out inducer-specific differential regulation of p49 and p56 both in vitro and in vivo. More impressive differential regulation was observed in B cells and pDCs. IFN-β or IFN-α could induce p56, p54, and p49 and their corresponding mRNAs equally well in T cells (Fig. 7 and data not shown). In contrast, p56 and its mRNA were poorly induced by IFN in B cells, although the other two members of the family were as strongly induced as in T cells. The possibility of a kinetic difference in gene induction was ruled out by performing a time course experiment (data not shown). Similarly, in mDCs all three proteins were induced by IFN but in pDCs, p56 was poorly induced (Fig. 8). These results demonstrate that in specific cells of the immune system IFN cannot induce p56, although the transcriptional signal is generated and two other closely related genes are strongly induced. To our knowledge, this is the first example of clearly cell-specific differential regulation of related ISGs.
In kidney mesangial cells, a different type of differential regulation was observed. In these cells, transfected dsRNA or SeV infection, both of which signal through the cytoplasmic receptors RIG-I/MDA-5, and IFN-α, could induce all three proteins efficiently. However, TLR3, activated by dsRNA added to the culture medium, or infection with VSV or EMCV could not induce any of the three proteins in mesangial cells, although in podocytes, which surround the mesangial cells in the glomeruli of the kidney, the latter inducers were very effective. We know that the mesangial cells express TLR3 and respond to added dsRNA by activating p38 and c-Src and we also know that VSV and EMCV can infect these cells (data not shown). Thus, the observed defects are probably not at the signal initiation stages but at the level of the lack of expression or functional defects in signaling proteins required further downstream of specific signaling pathways. Again, these defects are cell type specific and inducer specific.
Future investigation should explore the molecular basis for the different kinds of differential induction we documented here. Why is ISG56 not induced in B cells or pDCs, when the neighboring ISG54 and ISG49 are strongly induced? It is unlikely that any signaling defects or transcriptional inaccessibility of the ISG56 chromosomal region are responsible, because closely located genes were induced well. Determination of the mechanism of the observed ISG56-specific gene silencing will surely reveal unknown regulatory aspects of the IFN system. The mesangial cells revealed a different aspect of viral signaling pathways that lead to the induction of ISGs. Why were these cells, but not the podocytes, unresponsive to dsRNA, EMCV, and VSV? Observations of this kind question the validity of generalizing information about the specific pathway a virus uses to induce IFN and ISGs. It is not unlikely that in different cell types the same virus uses predominantly different signaling pathways. Indeed, our earlier studies, using mutant mice, showed that influenza A virus uses exclusively TLR pathways to induce IFN in pDCs, but in mDCs the cytoplasmic receptors are used exclusively (21). Similar scenarios may be true for other viruses, in other cell types. Studies, such as the one presented here, should increase our awareness of this additional level of complexity of the IFN system.
Acknowledgments
We thank Fulvia Terenzi for advice and for sharing reagents and Earl Poptic from the Lerner Research Institute Hybridoma Core for producing the p49 antibody.
This study was supported by National Institutes of Health grants CA068782 and CA062220.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413732-738. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J., S. VanScoy, T. F. Cheng, D. Gomez, and N. C. Reich. 2008. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9168-175. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bluyssen, H. A., R. J. Vlietstra, P. W. Faber, E. M. Smit, A. Hagemeijer, and J. Trapman. 1994. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics 24137-148. [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28655-662. [DOI] [PubMed] [Google Scholar]
- 6.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9515623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Veer, M. J., H. Sim, J. C. Whisstock, R. J. Devenish, and S. J. Ralph. 1998. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics 54267-277. [DOI] [PubMed] [Google Scholar]
- 8.Elco, C. P., J. M. Guenther, B. R. Williams, and G. C. Sen. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not Toll-like receptor 3. J. Virol. 793920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 10.Gilliet, M., A. Boonstra, C. Paturel, S. Antonenko, X. L. Xu, G. Trinchieri, A. O'Garra, and Y. J. Liu. 2002. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 195953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 765532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 196891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller, O., G. Kochs, and F. Weber. 2007. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 18425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershey, J. W., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 185-243. In N. Sonenberg, J. W. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press., Cold Spring Harbor, NY.
- 16.Hinnebusch, A. G. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31553-562. [DOI] [PubMed] [Google Scholar]
- 17.Hiscott, J. 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 28215325-15329. [DOI] [PubMed] [Google Scholar]
- 18.Hui, D. J., C. R. Bhasker, W. C. Merrick, and G. C. Sen. 2003. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J. Biol. Chem. 27839477-39482. [DOI] [PubMed] [Google Scholar]
- 19.Hui, D. J., F. Terenzi, W. C. Merrick, and G. C. Sen. 2005. Mouse p56 blocks a distinct function of eukaryotic initiation factor 3 in translation initiation. J. Biol. Chem. 2803433-3440. [DOI] [PubMed] [Google Scholar]
- 20.Kawai, T., and S. Akira. 2007. Antiviral signaling through pattern recognition receptors. J. Biochem. 141137-145. [DOI] [PubMed] [Google Scholar]
- 21.Kim, T. W., K. Staschke, K. Bulek, J. Yao, K. Peters, K. H. Oh, Y. Vandenburg, H. Xiao, W. Qian, T. Hamilton, B. Min, G. Sen, R. Gilmour, and X. Li. 2007. A critical role for IRAK4 kinase activity in Toll-like receptor-mediated innate immunity. J. Exp. Med. 2041025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi, N., N. Bagheri, J. G. Nedrud, R. M. Strieter, Y. Tomino, M. E. Lamm, and S. N. Emancipator. 2003. Differential effects of Sendai virus infection on mediator synthesis by mesangial cells from two mouse strains. Kidney Int. 641675-1684. [DOI] [PubMed] [Google Scholar]
- 23.Lee, C. G., J. Demarquoy, M. J. Jackson, and W. E. O'Brien. 1994. Molecular cloning and characterization of a murine LPS-inducible cDNA. J. Immunol. 1525758-5767. [PubMed] [Google Scholar]
- 24.Leonard, G. T., and G. C. Sen. 1996. Effects of adenovirus E1A protein on interferon-signaling. Virology 22425-33. [DOI] [PubMed] [Google Scholar]
- 25.Levy, D., A. Larner, A. Chaudhuri, L. E. Babiss, and J. E. Darnell, Jr. 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. USA 838929-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutz, M. B., N. Kukutsch, A. L. Ogilvie, S. Rossner, F. Koch, N. Romani, and G. Schuler. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 22377-92. [DOI] [PubMed] [Google Scholar]
- 27.McWhirter, S. M., K. A. Fitzgerald, J. Rosains, D. C. Rowe, D. T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 101233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84431-442. [DOI] [PubMed] [Google Scholar]
- 29.Mundel, P., J. Reiser, A. Zuniga Mejia Borja, H. Pavenstadt, G. R. Davidson, W. Kriz, and R. Zeller. 1997. Rearrangements of the cytoskeleton and cell contacts induce process formation during differentiation of conditionally immortalized mouse podocyte cell lines. Exp. Cell Res. 236248-258. [DOI] [PubMed] [Google Scholar]
- 30.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 2831150-1156. [DOI] [PubMed] [Google Scholar]
- 31.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5375-386. [DOI] [PubMed] [Google Scholar]
- 32.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 331027-1033. [DOI] [PubMed] [Google Scholar]
- 33.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito, T., and M. Gale, Jr. 2007. Principles of intracellular viral recognition. Curr. Opin. Immunol. 1917-23. [DOI] [PubMed] [Google Scholar]
- 35.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103245-259. [DOI] [PubMed] [Google Scholar]
- 37.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 3001148-1151. [DOI] [PubMed] [Google Scholar]
- 38.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 39.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 792689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terenzi, F., D. J. Hui, W. C. Merrick, and G. C. Sen. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 28134064-34071. [DOI] [PubMed] [Google Scholar]
- 41.Terenzi, F., S. Pal, and G. C. Sen. 2005. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology 340116-124. [DOI] [PubMed] [Google Scholar]
- 42.Terenzi, F., C. White, S. Pal, B. R. Williams, and G. C. Sen. 2007. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J. Virol. 818656-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wathelet, M., S. Moutschen, P. Defilippi, A. Cravador, M. Collet, G. Huez, and J. Content. 1986. Molecular cloning, full-length sequence and preliminary characterization of a 56-kDa protein induced by human interferons. Eur. J. Biochem. 15511-17. [DOI] [PubMed] [Google Scholar]
- 44.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 1752851-2858. [DOI] [PubMed] [Google Scholar]
- 45.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, Y., C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 8111246-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]