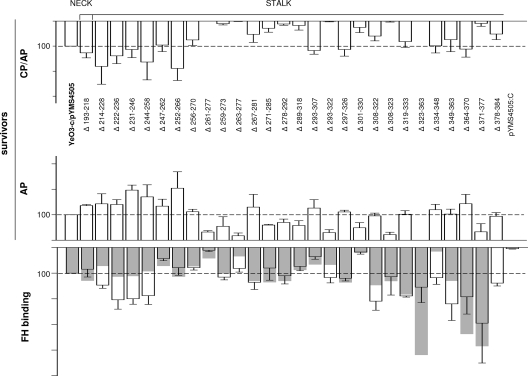
FIG. 3.
Deletion mapping of YadA for CR determinants. The resistance of the mutants to CP/AP- and AP-mediated killing is presented for the 2-h time point. For the same strains, the FH binding from HIS was determined by ELISA (for each strain, the transparent columns with standard deviation bars represent the average FH binding for three independent duplicate experiments) and by immunoblotting (superimposed gray columns) (quantitation of the FH band intensities was performed with the Typhoon 9400 Image Quant analyzer). Survival and FH binding values of the strain expressing wild-type YadA (YeO3-c/pYMS4505) were set to 100, and the survival percentage and FH binding of the mutants are expressed relative to those of the wild type. For the calculation of statistical significances between the deletion mutants and wild-type YadA, the actual survival percentages for each experiment were used. The average YeO3-c/pYMS4505 survival value for AP killing in serum pool 1 was 76.1 ± 22.1, and that in pool 2 was 103.7 ± 27.7; the value for the CP/AP killing in serum pool 1 was 49.4 ± 17.4, and that in pool 2 was 73.3 ± 7.2. In ELISA, the average optical density at 492 nm value for strain YeO3-c/pYMS4505 was 0.47 ± 0.19 (range, 0.27 to 0.92). The strain with a frameshift mutation in the yadA gene (YeO3-c/pYMS4505:C), unable to express YadA, was included as a negative control.