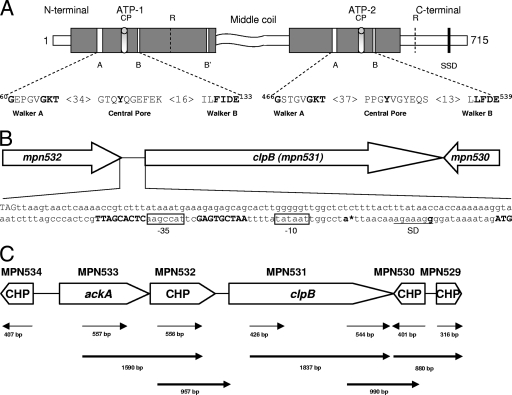
FIG. 1.
M. pneumoniae ClpB organization and transcription. (A) Proposed domain organization of M. pneumoniae ClpB and conserved motifs involved in ATP binding and ATP hydrolysis. M. pneumoniae ClpB retains only a shortened N-terminal domain, two nucleotide binding domains (ATP-1, positions 115 to 265; and ATP-2, positions 392 to 583) which are separated by the middle region, and a C-terminal region. Both nucleotide binding domains contain the Walker A (60GX4GKT69 and 466GX4GKT473) and Walker B (130Hy2DE133 and 536Hy2DE539) motifs, where Xn represents n amino acids of any kind and Hyn represents n hydrophobic amino acids. ATP-1 has one additional nucleotide binding motif, termed Walker B′ (249Hy2DE252). CPs (positions 102 to 111 and 511 to 519) contribute to protein substrate binding. Arg (R) residues (positions 187 and 609) are proposed to serve as Arg fingers, contacting ATP bound to an adjacent subunit. The sensor and substrate domain (SSD) (666GAR668) potentially senses the nucleotide status of ATP-2. The middle coiled-coil region is predicted to be involved in protein-protein interactions. (B) Promoter region of clpB. The bold ATG (far right) indicates the putative translational start site of clpB (mpn531). Inverted repeats on each side of the 9-nucleotide spacer sequence of the CIRCE element are shown in bold. The promoter region contains a Pribnow box (−10 site), which is an exact match for the consensus sequence (TATAAT) for the housekeeping sigma factor (σ70). A putative −35 site (boxed) was also identified upstream of the Pribnow box. The stop codon of mpn532 (TAG, far left) is 175 nucleotides upstream of the start codon of clpB. An asterisk designates the transcriptional start site (bold letter) as determined by PE. A Shine-Dalgarno (SD) sequence (underlined) is located downstream of the putative transcriptional start site. (C) Transcriptional analysis of genes surrounding clpB of M. pneumoniae. RT-PCR was performed on total RNA isolated from M. pneumoniae cells as described in Materials and Methods. A schematic of clpB and its surrounding genes is presented. Conserved hypothetical proteins (CHP) are indicated. Thin arrow lines represent regions that were amplified to confirm transcription of individual genes. Bold arrow lines represent RT-PCR products to indicate cotranscription of corresponding genes. Numbers represent the sizes of RT-PCR-amplified products.