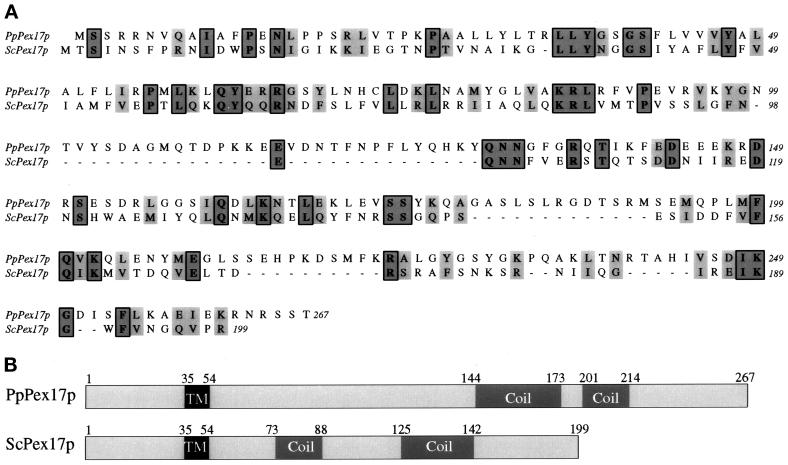
Figure 3.
Sequence alignment and features of P. pastoris and S. cerevisiae Pex17p orthologues. The amino acid sequences were aligned using the ClustalW program (A). Identical residues (boxed) and similar residues (gray) are shaded. Similarity rules: G = A = S, A = V, V = I = L = M, I = L = M = F = Y = W, K = R = H, D = E = Q = N, and S = T = Q = N. Dashes represent gaps. (B) Sequence features and relative positions. TM, putative transmembrane domain; Coil, putative coiled-coil domain.