Abstract
Dominant disease alleles are attractive therapeutic targets for allele-specific gene silencing by small interfering RNA (siRNA). Sialuria is a dominant disorder caused by missense mutations in the allosteric site of GNE, coding for the rate-limiting enzyme of sialic acid biosynthesis, UDP-GlcNAc 2-epimerase/ManNAc kinase. The resultant loss of feedback inhibition of GNE-epimerase activity by CMP-sialic acid causes excessive production of free sialic acid. For this study we employed synthetic siRNAs specifically targeting the dominant GNE mutation c.797G>A (p.R266Q) in sialuria fibroblasts. We demonstrated successful siRNA-mediated down-regulation of the mutant allele by allele-specific real-time PCR. Importantly, mutant allele-specific silencing resulted in a significant decrease of free sialic acid, to within the normal range. Feedback inhibition of GNE-epimerase activity by CMP-sialic acid recovered after silencing demonstrating specificity of this effect. These findings indicate that allele-specific silencing of a mutated allele is a viable therapeutic strategy for autosomal dominant diseases, including sialuria.—Klootwijk, R. D., Savelkoul, P. J. M., Ciccone, C., Manoli, I., Caplen, N. J., Krasnewich, D. M., Gahl, W. A., Huizing, M. Allele-specific silencing of the dominant disease allele in sialuria by RNA interference.
Keywords: real-time PCR, feedback inhibition, sialic acid, UDP-GlcNAc 2-epimerase/ManNAc kinase, siRNA, allosteric site
Sialuria (mim 269921) is a rare, autosomal dominant inborn error of metabolism in which excessive free N-acetylneuraminic acid (Neu5Ac, henceforth referred to as “sialic acid”) is synthesized in the cellular cytoplasm. Sialic acids are typically found as the terminal sugars on glycoconjugates, where they serve a variety of cellular signaling functions (1, 2). Sialuria patients manifest mild coarse facies and slight motor delay, with additional sporadic features of hepatosplenomegaly, delayed skeletal development, microcytic anemia, and mild intellectual impairment (3,4,5). They are diagnosed by the detection of gram quantities of free sialic acid in urine and significantly increased concentrations of free sialic acid in the cytoplasm of cultured fibroblasts (5,6,7).
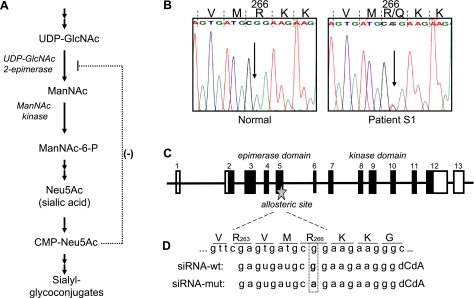
The biochemical defect of sialuria involves failure to regulate sialic acid synthesis, due to impaired allosteric feedback inhibition of the rate-limiting, bifunctional enzyme uridine diphospho-N-acetylglucosamine (UDP-GlcNAc) 2-epimerase/N-acetylmannosamine (ManNAc) kinase (GNE/MNK), by cytidine monophosphate (CMP) -sialic acid (Fig. 1A) (6, 8,9,10,11). One mutation in the allosteric domain of GNE, coding for the GNE/MNK protein, impairs feedback inhibition and results in excessive production of cytoplasmic sialic acid. In fact, all known sialuria patients are heterozygous for a missense mutation in 1 of 2 codons of the GNE gene, codon 263 (R263L) or codon 266 (mutations R266Q and R266W), marking the region of codons 263–266 as the allosteric site for CMP-sialic acid binding (10, 12). However, the full extent of the allosteric site has not been formally defined.
Figure 1.
Intracellular sialic acid synthesis, sialuria mutation, GNE gene structure, and design of allele-specific siRNA. A) The synthesis of sialic acid is initiated in the cytosol, where glucose undergoes several modifications to eventually become Neu5Ac (sialic acid). The reactions catalyzed by the UDP-GlcNAc 2-epimerase (GNE)/ManNAc kinase (MNK) bifunctional enzyme are the central and rate-limiting steps in this cytosolic process. GNE-epimerase activity is feedback inhibited by the downstream product CMP-Neu5Ac (CMP-sialic acid). CMP-sialic acid is utilized by the Golgi complex to produce sialyl glycoconjugates. B) The heterozygous GNE mutation, c.797G>A, in sialuria results in a R266Q missense amino acid change at the protein level. This mutation is located in the allosteric site of the GNE-epimerase enzymatic domain. C) Schematic of the GNE gene structure (not to scale). GNE consists of 13 exons, coding for the 722-aa protein GNE/MNK. The N-terminal part (exons 2–6) codes for the GNE-epimerase catalytic domain, including its allosteric site. The C-terminal part (exons 7–12) codes for the ManNAc-kinase catalytic domain. D) Schematic of the allosteric site of GNE/MNK, defined by amino acids R263 and R266, and design of the allele-specific siRNAs employed in this study.
Dominant human disorders due to failed allosteric inhibition are extremely rare (13,14,15), and no specific therapies are available. However, recent in vitro studies have shown that allele-specific silencing of dominant disease genes through exploitation of the naturally occurring gene-silencing mechanism RNA interference (RNAi) might be beneficial for this group of disorders (16,17,18,19). Sialuria cells could serve as a model to demonstrate the efficacy of this technique.
In this study, we used RNAi mediated by synthetic small interfering RNAs (siRNAs) to induce allele-specific silencing of a mutant GNE c.797G>A transcript (p.R266Q; Fig. 1B) in primary fibroblasts from a sialuria patient. Allele-specific silencing was confirmed at the RNA level employing a semiquantitative allele-specific real-time PCR method, which we describe in this article. In addition, allele-specific silenced cells showed significantly decreased free sialic acid levels and restoration of enzymatic feedback inhibition by CMP-sialic acid. These findings demonstrate proof of principle for the therapeutic potential of allele-specific silencing for sialuria and other dominantly inherited disorders.
MATERIALS AND METHODS
Mutation analysis
For GNE mutation analyses, genomic DNA was PCR-amplified exon-by-exon and directly sequenced (primers and conditions available on request). The GNE allosteric site in exon 5, mutated in sialuria, was amplified with the forward primer 5′-TGAGTTCCTAGATGAGTGAAG-3′ and reverse primer 5′-CAGGTTGATCACAGGTGTT-3′. Automated sequencing was performed on a Beckman CEQ 2000, using the CEQ Dye Terminator Cycle Sequencing kit according to the manufacturer’s protocols (Beckman Coulter, Fullerton, CA, USA).
siRNA design
Allele-specific siRNAs manufactured for these studies (Qiagen, Valencia, CA, USA) included the following (see also Fig. 1D): 1) siRNA against the sialuria patient mutation c.797G>A (siRNA-mut): sense 5′ GAGUGAUGCAGAAGAAGGGdCdA 3′, antisense 5′ CCCUUCUUCUGCAUCACUCdGdA 3′; 2) siRNA directed against the wild-type allele (siRNA-wt): sense 5′ GAGUGAUGCGGAAGAAGGGdCdA 3′, antisense 5′ CCCUUCUUCCGCAUCACUCdGdA 3′; 3) Control siRNA (siRNA-nonsil): sense 5′ UUCUCCGAACGUGUCACGUdTdT 3′, antisense 5′ UCGUGACACGUUCGGAGAAdTdT 3′ (siRNA-nonsil). The siRNA-nonsil had no bioinformatically predicted sequence target in the human genome and was used as a negative control (AllStars Negative Control siRNA, Qiagen) for real-time PCR analyses and GNE-epimerase enzyme activity assays to assess the specificity of the siRNA-mut and siRNA-wt.
Cell culture and transfections
Sialuria fibroblasts, kindly provided by Dr. Clara Sá Miranda (Instituto de Genetica Medica, Porto, Portugal), and normal fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum and 100 mg/ml of primocin (Invivogen, San Diego, CA, USA) at 37°C in 5% CO2. Fibroblasts were transfected using the human dermal fibroblast nucleofector kit according to the manufacturer’s protocol (Amaxa Biosystems, Gaithersburg, MD, USA). Briefly, 5 × 105 cells were transfected in 100 μl nucleofector solution with 1.5 μg siRNA and grown in 6 well plates for 48 h. Next, cells were washed with ice-cold PBS, scraped in 1.5 ml cold PBS on ice, and centrifuged for 5 min at 900 g. Cell pellets were resuspended in 60 μl H2O (Nanopure Diamond water purification systems; Barnstead International, Dubuque, IA, USA; 18.2 MÙ · cm) and sonicated in a Microson (Microsonix Inc., New York, NY, USA). The cell suspensions were centrifuged for 15 min at 20,800 g, and the supernatants were used for sialic acid measurements and enzymatic assays. Aliquots of the supernatants were analyzed for total protein concentration using the bicinchoninic acid (BCA) protein assay kit according to the manufacturer’s protocol (Pierce Biotechnology, Rockford, IL, USA).
In vitro transcription
The human GNE coding sequence (GenBank NM_005476) was amplified from cDNA of normal human fibroblasts and subcloned, using EcoRI and XhoI restriction sites, into the pET17b vector (EMD Biosciences, San Diego, CA, USA), which contained an N-terminal T7 tag and a T7 promotor sequence. This GNE-pET17b plasmid was used to create the patient-specific c.797G>A GNE target mutation by site-directed mutagenesis using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). All constructs were sequence verified before use. PCR amplifications of the GNE coding sequence, including the T7 promotor sequence of pET17b, were performed using pure Taq Ready-To-Go PCR beads (Amersham Biosciences, Piscataway, NJ, USA). Amplifications were performed in a 25 μl reaction volume containing 25 ng GNE-pET17b plasmid DNA (normal or mutated) and 0.4 μM of each primer flanking the T7 promotor (forward, 5′-TAATACGACTCACTATAGGG-3′) and the GNE termination codon (reverse, 5′-GCTAGTTATTGCTCAGCGG-3′). After amplification, the T7 promotor sequence on each PCR fragment was employed for RNA transcription of normal and mutated GNE using the MAXIscript In Vitro Transcription Kit according to the manufacturer’s protocol (Ambion, Austin, TX, USA).
DNA, cDNA, and RNA preparation
Skin fibroblasts of patients and healthy controls were plated and grown to 80–90% confluency, after which genomic DNA or total RNA was isolated. Genomic DNA was obtained using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA). Total RNA was isolated using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). Bacterial total RNA was isolated from TOP10 chemically competent Escherichia coli cells (Invitrogen) using the Trizol Max Bacterial RNA isolation kit (Invitrogen). All RNA samples were treated with RNase-free DNase (rDNAse; Ambion), and purified using RNeasy mini columns (Qiagen). First strand cDNA synthesis was performed using the High Capacity cDNA Archive kit employing oligo-dT primers (Applied Biosystems, Foster City, CA, USA), after which the cDNA was purified using RNeasy minicolumns (Qiagen). Concentrations of RNA, genomic DNA, and cDNA were measured with a GeneQuant Pro spectrophotometer (Amersham Biosciences).
Real-time PCR
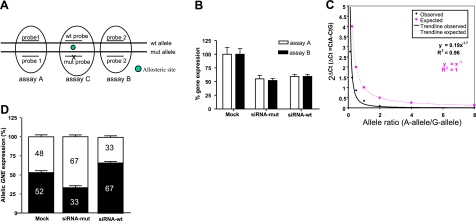
Quantitative real-time PCR was performed on RNA isolated from sialuria patient fibroblasts, utilizing assays-on-demand (Applied Biosystems) for GNE [Fig. 2A: assay A, Hs01103397_m1 (GNE exon 4–5), and assay B, Hs01103400_m1, (GNE exon 5–6)] and TATA Box Binding Protein (Hs00427620_m1, control probe), on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). Gene expression levels were determined using the ΔΔCt method (20). To measure allelic RNA expression levels, we developed a semiquantitative allele-specific RNA assay, based on the method described by Suda et al. (21). For this assay, the region flanking the GNE target mutation was PCR amplified using a specific primer set (5′-ATTGACGCAGGGAGCAAAGA-3, 5′-GGATGATGCTCAATGCCCTTCTT-3′) in the presence of 2 probes, each selectively hybridizing to the normal (5′ VIC-labeled, 5′-NFQ-CATCACTCGAACCATC-VIC-3′) or the mutated (5′ FAM-labeled, 5′-NFQ-CATCACTAGAACCATC-FAM-3′) allele (manufactured by Applied Biosystems). For each allele-specific real-time PCR reaction, a total reaction mixture of 25 μl contained 900 nM primers and 200 nM allele-specific VIC/FAM probes, 50 ng of cDNA, and Taqman Universal PCR Master Mix (Applied Biosystems). All real-time PCR reactions and subsequent analyses were performed on an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The prerun thermal cycling conditions were 10 min at 95°C to activate the TaqDNA-polymerase, followed by 40 cycles of 95°C for 15 s and 60°C annealing/extension for 1 min. Each experiment was performed in triplicate. Within each experiment, reactions were run in duplicate. For the target GNE mutation, a standard curve was constructed (Fig. 2C) by mixing the normal and mutant cDNA samples (created by in vitro transcription of normal and mutated T7-GNE DNA, described above) to allelic ratios of 8, 4, 2, 1, 0.5, 0.25, and 0.125 in a total concentration of 50 ng/μl. In addition, 10 ng/μl bacterial cDNA was added to each standard curve reaction mixture to mimic nonspecific cDNA present in the fibroblast samples.
Figure 2.
Allele-specific silencing of GNE in sialuria fibroblasts by siRNA. A) Location of the primer-probe assays used for gene expression analysis by real-time PCR in the GNE gene. Assays A and B, located over GNE exon-exon boundaries (exon 4–5 and exon 5–6, respectively), were employed for determination of total GNE expression. Assay C, located in exon 5 over the GNE mutation of the sialuria patient, was used for allele-specific expression analysis of the mutated (c.797A) and the wild-type (c.797G) allele. B) Real-time PCR analysis of total GNE RNA expression in sialuria fibroblasts 48 h after mock (no siRNA), siRNA-mut, and siRNA-wt transfections. C) Standard curve produced by mixing allelic quantities of mutated allele A vs. wild-type allele G, which was then used for determination of allelic expressions in patient cells. D) Allelic expressions of GNE [wild-type G-allele (white) vs. mutant A-allele (black)] in sialuria cells 48 h after mock (no siRNA), si-RNA-mut, and siRNA-wt transfections.
GNE enzyme assay
For GNE-epimerase enzymatic measurements the conversion of UDP-[3H]GlcNAc to [3H]ManNAc was assayed as described (8, 22). Briefly, in a final reaction volume of 117.5 μl, 5 μl UDP-[3H]GlcNAc (1 mCi/ml, 60 Ci/mmol) (American Radiolabeled Chemicals, St. Louis, MO, USA) was added to a mixture containing 64 μM UDP-GlcNAc, 10 mM MgCl2, and 75 μl of protein/enzyme mixture. The protein/enzyme mixture consisted of fibroblasts lysed in buffer A (pH 7.2, containing 0.2 M MES, 0.15 M NaCl, 10 mM CaCl2, 1% Triton X-100, 4 μg/ml aprotinin, and 0.1 mM UDP). The total protein concentration in the fibroblast lysates was determined using the BCA Protein Assay Reagent Kit (Pierce Biotechnology). The enzymatic activity of GNE-epimerase was measured in the absence or presence of 100 μM CMP-sialic acid (to determine feedback inhibition). The GNE-epimerase reactions were performed by incubation for 6 min at 37°C, and stopped by boiling the sample for 1 min. After incubation on ice, the samples were centrifuged at 13,000 g for 15 min at 4°C. The supernatants were removed and frozen at −20°C until chromatographic separation by high-pH anion exchange chromatography on a BioLC carbohydrate analyzer (Dionex, Sunnyvale, CA, USA) and subsequent [3H]ManNAc analysis.
HPLC analyses of sialic acid and ManNAc
We employed the BioLC carbohydrate analyzer (Dionex), which uses high-pH anion-exchange chromatography with pulsed amperometric detection to separate monosaccharides based on charge as well as size and composition. For GNE-epimerase assays, 30 μl of each supernatant, which had undergone the enzyme assay, was applied to a CarboPac PA-100 column, after which UDP-GlcNAc, GlcNAc, and ManNAc were separated by using an isocratic solution of 10 mM NaOH followed by an increasing (0–900 mM) NaOAc gradient. Here 20 μl of GlcNAc/ManNAc (for the GNE-epimerase assay) was coinjected to identify migration times of the monosaccharides. Fractions were collected at 1 min intervals. Eluted fractions were measured for [3H] radioactivity using a MicroBeta Scintillation Counter (Perkin Elmer, Shelton, CT, USA), and were converted into picomoles based on the specific radioactivity of the substrate. For sialic acid measurements, 50 μl of each fibroblast supernatant was applied to a CarboPac PA-100 column, after which free sialic acid was separated by eluting with an isocratic solution of 100 mM NaOH. Our methodology measured only free sialic acid, not CMP-sialic acid. CMP-sialic acid is more charged than free sialic acid and requires a higher acetate concentration for elution from the column.
Statistical analysis
The 2-tailed unpaired t test was used for statistical analysis of data obtained by real-time PCR assays, sialic acid measurements, and feedback inhibition experiments.
RESULTS
Clinical features of sialuria
The sialuria fibroblasts were from a 14-yr-old Portuguese female previously described (7). Urinary excretion of free sialic acid was 19 μmol sialic acid/mg creatinine (normal, 0.24–0.64). The cultured fibroblasts contained 17.6 nmol sialic acid/mg protein (normal, 0.2–3.3). The patient was heterozygous for the GNE missense mutation c.797G>A (p.R266Q), located within the GNE/MNK allosteric site (Fig. 1B).
Silencing of GNE RNA
Synthetic siRNA oligonucleotides were designed to suppress selectively mutant GNE A-allele (siRNA-mut), complementary to the c.797G>A mutation in the allosteric site of GNE, and to suppress selectively the wild-type GNE G-allele (siRNA-wt) (Fig. 1C). An siRNA (siRNA-nonsil) with no bioinformatically predicted sequence target in the human genome was used as a negative control.
The silencing mediated by each of the synthetic GNE siRNAs was first assessed by analyzing total GNE RNA levels (mutated and wild-type alleles combined) in sialuria fibroblasts, 48 h posttransfection, using real-time quantitative PCR analysis with primer-probes located upstream (assay A) and downstream (assay B) of the GNE allosteric site (Fig. 2A). Transfection with siRNA-mut resulted in a 1.9-fold decrease in total GNE expression (assay A, P=0.02; assay B, P=0.03) (Fig. 2B). A comparable 1.7-fold decrease in total GNE RNA expression was measured after transfection with siRNA-wt (assay A, P=0.02; assay B, P=0.04) (Fig. 2B). No decrease in total GNE expression was observed using the control siRNA-nonsil (not shown).
Next, we developed a semiquantitative allele-specific RNA assay to quantify allele-specific silencing. This method is based on the presence of 2 allele-specific probes in 1 real-time PCR assay (assay C in Fig. 2A); each probe specifically recognizes and quantifies 1 of the alleles. A standard curve for this assay was constructed by mixing known allelic ratios of normal and mutant cDNA samples, created by in vitro transcription of RNA from normal and mutated T7-GNE DNA (Fig. 2C). The RNA allele-ratio for each test sample could then be determined using the standard curve. RNA isolated from mock (no siRNA) transfected sialuria fibroblasts yielded an allelic ratio of 1.1 (1.0–1.2; n=9) (Fig, 2C, D), indicating that the A-allele and the G-allele at position 797 of GNE mRNA in this patient are expressed in similar amounts, as expected. Transfection of sialuria fibroblasts with the siRNA-mut synthetic siRNA resulted in a significant decrease in the allelic ratio of mutant A-allele/wild-type G-allele, from 1.1 to 0.5 (0.4–0.5; n=8) (Fig. 2D). Transfection with the siRNA-wt synthetic siRNA resulted in a significant increase in the ratio of mutant A-allele/wild-type G-allele, from 1.1 to 2.0 (1.8–2.2; n=9) (Fig. 2D). These PCR analyses demonstrate that the siRNA-mut and siRNA-wt synthetic siRNAs each significantly silenced their target alleles, with some nonspecific silencing of the other (nontargeted) allele.
Cytoplasmic sialic acid levels after siRNA silencing
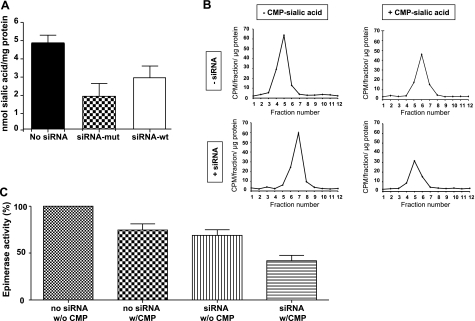
The free sialic acid content of sialuria fibroblasts was determined before and after siRNA transfection. In transfected cells, siRNA-mut induced significant reduction of free sialic acid, to 35 ± 11% (n=3; P=0.01), compared to mock (no siRNA)-transfected cells (Fig. 3A). Transfection with siRNA-wt also caused a small, but not statistically significant, decrease in free sialic acid to 60 ± 13% (n=3; P=0.07) compared to mock-transfected cells (Fig. 3A), indicating that siRNA-wt exhibited some nonspecific silencing of the mutant allele.
Figure 3.
Free sialic acid levels and GNE-epimerase activity in sialuria fibroblasts before and after siRNA-mediated silencing. A) Free sialic acid levels in sialuria fibroblasts 48 h after mock (no siRNA), siRNA-mut, and siRNA-wt transfections. B) GNE-epimerase activity and inhibition with and without siRNA transfection. UDP-[3H]GlcNAc was added as substrate to extracts of nonsilenced (−siRNA) and siRNA-mut silenced (+siRNA) sialuria fibroblasts in the presence (+) and absence (−) of 100 μM CMP-sialic acid. On separation of the reaction products, [3H]ManNAc (CPM/fraction/μg protein) was determined in each fraction and served as a measure of GNE-epimerase enzymatic activity. C) GNE-epimerase activity determined from the graphs in B. Enzyme activities in the presence (w/CMP) or absence (w/o CMP) of 100 μM CMP-sialic acid in mock (no siRNA) or siRNA-mut (siRNA) transfected cells were calculated relative to the average GNE-epimerase activity of mock transfected cells (no siRNA w/o CMP), which was set to 100%.
GNE enzymatic feedback inhibition after silencing
In sialuria the product of the mutant GNE allele, GNE-epimerase, fails to be feedback inhibited by CMP-sialic acid (6, 8, 10, 12). We determined GNE-epimerase enzyme activity in sialuria fibroblast extracts, and the enzyme’s sensitivity to feedback inhibition by 100 μM CMP-sialic acid, before and after silencing of the mutant allele. Epimerase activity (8, 22) was set to 100% in fibroblasts of patient S1 in the absence of CMP-sialic acid. In the presence of CMP-sialic acid, the sialuria fibroblasts showed only a slight (∼25%; n=3) reduction in GNE-epimerase activity (Fig. 3B, C), indicating defective feedback inhibition. Published levels of inhibition of GNE-epimerase activity by 100 μM CMP-sialic acid are 79–100% for normal fibroblasts and 19–27% for sialuria cells (6, 7). Transfection with siRNA-mut reduced the GNE-epimerase enzyme activity by ∼30% (n=2), reflecting reduced translation of the GNE/MNK enzyme. The reduced level of GNE-epimerase activity was lowered an additional 40% in presence of CMP-sialic acid (Fig. 3B, C; n=3, P=0.05).
DISCUSSION
Gene silencing mediated through the RNAi pathway by siRNAs has the potential to treat diseases caused by dominant, gain-of-function mutations. A major challenge is to specifically inhibit the disease-causing allele while maintaining function of the normal allele.
The disorder sialuria, for which only symptomatic therapy is available, provides an excellent example of a disease caused by a dominant missense mutation. We examined the effects of siRNA-mediated silencing of the mutant allele in sialuria, relying on the remaining normal allele to provide regulated GNE-epimerase enzymatic activity for sialic acid synthesis.
Real-time PCR showed that silencing of the mutated allele c.797G>A (p.R266Q) in sialuria fibroblasts reduced total GNE RNA levels by ∼50% (Fig. 2B), suggesting that expression of the normal allele was untouched. To verify this, we developed a novel allele-specific real-time PCR assay, which demonstrated that the mutant allele was greatly silenced, but some residual mutant RNA (33%) remained present (Fig. 2D).
Critically, the end result was a reduction of free sialic acid levels in sialuria fibroblasts to ∼2 nmol/mg protein (Fig. 3A), which is within the normal range (0.2–3.3 nmol/mg protein) (6, 7, 23). The amount of free sialic acid (not CMP-sialic acid) is a good measure of impaired feedback inhibition of GNE-epimerase activity in sialuria fibroblasts. This is because free sialic acid is hugely overproduced (4–273 nmol/mg protein; normal, 0.2–3.3 nmol/mg protein) (6, 7, 23), while CMP-sialic acid levels remain modest (2–5 nmol/mg protein) in sialuria fibroblasts (6).
Silencing of the mutant GNE allele also reduced GNE-epimerase activity to ∼60% of that in nonsilenced cells (Fig. 3C) and restored GNE-epimerase feedback inhibition by 100 μM CMP-sialic acid to 60% (Fig. 3C), close to that of normal fibroblasts, 79–100% (6, 7). Residual mutant GNE/MNK enzyme, present after silencing (Fig. 2D), was the likely source of enzyme that was resistant to feedback inhibition.
Besides the sialuria mutations, other examples of dominant disease alleles being silenced for therapeutic reasons include mutations in the rhodopsin gene in retinitis pigmentosa (24), the Cu, Zn superoxide dismutase gene in amyotrophic lateral sclerosis (25,26,27), the torsinA encoding gene in DYT1 dystonia (28), acetylcholine receptor subunits in slow channel congenital myasthenic syndrome (16), and fibroblast growth factor receptor 2 in Apert syndrome (29). None of these other diseases, however, involves defective allosteric enzyme inhibition. Our demonstration of successfully silencing the dominant GNE mutation in sialuria should prompt consideration of similar silencing approaches for dominant allosteric enzyme defects. Such disorders include hyperinsulinism-hyperammonemia syndrome (MIM 606762) caused by allosteric mutations in the glutamate dehydrogenase gene (13, 14) and phosphoribosylpyrophosphate synthetase I superactivity (X-linked, MIM 300661), in which an allosteric site mutation results in loss of purine nucleotide feedback inhibition (15, 30).
In fact, any disorders caused by dominant gain-of-function mutations are credible candidates for allele-specific silencing-based therapeutics (17). Examples are abundant and include constitutive activation of G-protein-coupled receptors, several connective tissue diseases, progressive neurodegenerative disorders, and cancers. Mutations causing aberrant splice forms (exon-skipping) are also suitable targets for siRNA-based therapeutic stategies (31).
Although siRNA-based therapies are promising, their risks require further investigation (17, 31, 32). The techniques involved have been utilized only in preclinical testing using animal models of different diseases (32,33,34). Moreover, sialuria and other dominant disorders manifest systemic complications requiring siRNA targeting of several tissues, including the central nervous system. Hence, the first applications of synthetic siRNA therapeutics may occur in disorders in which the affected organs are easily accessible using gene-therapy based methods, such as the eye (24, 35), lung (36, 37), or skin (38, 39). For these disorders, it may be also feasible to identify specific inhibitors that achieve allele-specific silencing and offer therapeutic promise. Such inhibitors may be identified through protein-epitope specific antibody development or by use of rapidly developing chemical genomics screening techniques (40, 41).
For many disorders, mutational heterogeneity presents a significant barrier to development of allele-based therapies. In contrast, sialuria is caused by GNE mutations affecting only 1 of 2 amino acids and serves as a good candidate for allele-based silencing. Very few sialiuria patients have been reported (3,4,5, 7, 10,11,12), but the prevalence is probably underestimated because of the mildness of the disorder. In addition, the assay of urinary free sialic acid is not a routine laboratory procedure. For individuals with mild developmental delay, we suggest consideration of this clinical measurement. Finally, should reasonable therapy emerge for sialic acid storage disorders, free sialic acid measurements could be added as part of routine newborn screening. Such screening would not only diagnose patients with sialuria, but also with Salla disease (MIM 604369) and infantile free sialic acid storage disorder (ISSD, MIM 269920), both caused by recessive mutations in the lysosomal sialic acid transporter SLC17A5 (42, 43).
Acknowledgments
This study was supported by the Intramural Research programs of the National Human Genome Research Institute and the National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
References
- Varki A. Sialic acids as ligands in recognition phenomena. FASEB J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- Varki A. Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature. 2007;446:1023–1029. doi: 10.1038/nature05816. [DOI] [PubMed] [Google Scholar]
- Montreuil J, Biserte G, Strecker G, Spik G, Fontaine G, Farriaux J P. Description of a new type of melituria: sialuria. C R Acad Sci Hebd Seances Acad Sci D. 1967;265:97–99. [PubMed] [Google Scholar]
- Krasnewich D M, Tietze F, Krause W, Pretzlaff R, Wenger D A, Diwadkar V, Gahl W A. Clinical and biochemical studies in an American child with sialuria. Biochem Med Metab Biol. 1993;49:90–96. doi: 10.1006/bmmb.1993.1010. [DOI] [PubMed] [Google Scholar]
- Enns G M, Seppala R, Musci T J, Weisiger K, Ferrell L D, Wenger D A, Gahl W A, Packman S. Clinical course and biochemistry of sialuria. J Inherit Metab Dis. 2001;24:328–336. doi: 10.1023/a:1010588115479. [DOI] [PubMed] [Google Scholar]
- Seppala R, Tietze F, Krasnewich D, Weiss P, Ashwell G, Barsh G, Thomas G H, Packman S, Gahl W A. Sialic acid metabolism in sialuria fibroblasts. J Biol Chem. 1991;266:7456–7461. [PubMed] [Google Scholar]
- Ferreira H, Seppala R, Pinto R, Huizing M, Martins E, Braga A C, Gomes L, Krasnewich D M, Sa Miranda M C, Gahl W A. Sialuria in a Portuguese girl: clinical, biochemical, and molecular characteristics. Mol Genet Metab. 1999;67:131–137. doi: 10.1006/mgme.1999.2852. [DOI] [PubMed] [Google Scholar]
- Weiss P, Tietze F, Gahl W A, Seppala R, Ashwell G. Identification of the metabolic defect in sialuria. J Biol Chem. 1989;264:17635–17636. [PubMed] [Google Scholar]
- Hinderlich S, Stäsche R, Zeitler R, Reutter W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver: purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- Seppala R, Lehto V P, Gahl W A. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am J Hum Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M. Disease mechanisms associated with mutations of the GNE gene. Drug Disc Today. 2005;2:519–527. [Google Scholar]
- Leroy J G, Seppala R, Huizing M, Dacremont G, De Simpel H, Van Coster R N, Orvisky E, Krasnewich D M, Gahl W A. Dominant inheritance of sialuria, an inborn error of feedback inhibition. Am J Hum Genet. 2001;68:1419–1427. doi: 10.1086/320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley C A, Lieu Y K, Hsu B Y, Burlina A B, Greenberg C R, Hopwood N J, Perlman K, Rich B H, Zammarchi E, Poncz M. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med. 1998;338:1352–1357. doi: 10.1056/NEJM199805073381904. [DOI] [PubMed] [Google Scholar]
- Fang J, Hsu B Y, MacMullen C M, Poncz M, Smith T J, Stanley C A. Expression, purification and characterization of human glutamate dehydrogenase (GDH) allosteric regulatory mutations. Biochem J. 2002;363:81–87. doi: 10.1042/0264-6021:3630081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M A, Taylor W, Smith P R, Ahmed M. Overexpression of the normal phosphoribosylpyrophosphate synthetase 1 isoform underlies catalytic superactivity of human phosphoribosylpyrophosphate synthetase. J Biol Chem. 1996;271:19894–19899. doi: 10.1074/jbc.271.33.19894. [DOI] [PubMed] [Google Scholar]
- Abdelgany A, Wood M, Beeson D. Allele-specific silencing of a pathogenic mutant acetylcholine receptor subunit by RNA interference. Hum Mol Genet. 2003;12:2637–2644. doi: 10.1093/hmg/ddg280. [DOI] [PubMed] [Google Scholar]
- Miller V M, Xia H, Marrs G L, Gouvion C M, Lee G, Davidson B L, Paulson H L. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci U S A. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Yokota T, Mitani T, Ito K, Anzai M, Miyagishi M, Taira K, Mizusawa H. Transgenic small interfering RNA halts amyotrophic lateral sclerosis in a mouse model. J Biol Chem. 2005;280:42826–42830. doi: 10.1074/jbc.M507685200. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Paulson H L. Allele-specific RNA interference for neurological disease. Gene Ther. 2006;13:576–581. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- Livak K J. Foster City, CA, USA: PE Applied Biosystems; Comparative Ct method. ABI Prism 7700 Sequence Detection System. User Bulletin 2. 1997 [Google Scholar]
- Suda T, Katoh M, Hiratsuka M, Fujiwara M, Irizawa Y, Oshimura M. Use of real-time RT-PCR for the detection of allelic expression of an imprinted gene. Int J Mol Med. 2003;12:243–246. [PubMed] [Google Scholar]
- Sparks S E, Ciccone C, Lalor M, Orvisky E, Klootwijk R, Savelkoul P J, Dalakas M C, Krasnewich D M, Gahl W A, Huizing M. Use of a cell-free system to determine UDP-N-acetylglucosamine 2-epimerase and N-acetylmannosamine kinase activities in human hereditary inclusion body myopathy. Glycobiology. 2005;15:1102–1110. doi: 10.1093/glycob/cwi100. [DOI] [PubMed] [Google Scholar]
- Lemyre E, Russo P, Melancon S B, Gagne R, Potier M, Lambert M. Clinical spectrum of infantile free sialic acid storage disease. Am J Med Genet. 1999;82:385–391. [PubMed] [Google Scholar]
- O'Reilly M, Palfi A, Chadderton N, Millington-Ward S, Ader M, Cronin T, Tuohy T, Auricchio A, Hildinger M, Tivnan A, McNally N, Humphries M M, Kiang A S, Humphries P, Farrar G J. RNA interference-mediated suppression and replacement of human rhodopsin in vivo. Am J Hum Genet. 2007;81:127–135. doi: 10.1086/519025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Schwarz D S, Keene A, el Affar B, Fenton L, Xia X, Shi Y, Zamore P D, Xu Z. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- Maxwell M M, Pasinelli P, Kazantsev A G, Brown R H., Jr RNA interference-mediated silencing of mutant superoxide dismutase rescues cyclosporin A-induced death in cultured neuroblastoma cells. Proc Natl Acad Sci U S A. 2004;101:3178–3183. doi: 10.1073/pnas.0308726100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Zhou H, Huang Y, Xu Z. Allele-specific RNAi selectively silences mutant SOD1 and achieves significant therapeutic benefit in vivo. Neurobiol Dis. 2006;23:578–586. doi: 10.1016/j.nbd.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Bode N, Davidson B L, Paulson H L. Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J Neurosci. 2005;25:10502–10509. doi: 10.1523/JNEUROSCI.3016-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V, Coumoul X, Wang R H, Kim H S, Deng C X. RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nat Genet. 2007;39:1145–1150. doi: 10.1038/ng2096. [DOI] [PubMed] [Google Scholar]
- Becker M A, Smith P R, Taylor W, Mustafi R, Switzer R L. The genetic and functional basis of purine nucleotide feedback-resistant phosphoribosylpyrophosphate synthetase superactivity. J Clin Invest. 1995;96:2133–2141. doi: 10.1172/JCI118267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, van Ommen G J. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Kay M A. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? J Clin Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson H L, Davidson B L. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lebron E, Denovan-Wright E M, Nash K, Lewin A S, Mandel R J. Intrastriatal rAAV-mediated delivery of antihuntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman S M, Binkley E A, Kumar-Singh R. Towards mutation-independent silencing of genes involved in retinal degeneration by RNA interference. Gene Ther. 2005;12:1223–1228. doi: 10.1038/sj.gt.3302512. [DOI] [PubMed] [Google Scholar]
- Birchall J. Pulmonary delivery of nucleic acids. Expert Opin Drug Deliv. 2007;4:575–578. doi: 10.1517/17425247.4.6.575. [DOI] [PubMed] [Google Scholar]
- Massaro D, Massaro G D, Clerch L B. Noninvasive delivery of small inhibitory RNA and other reagents to pulmonary alveoli in mice. Am J Physiol. 2004;287:L1066–L1070. doi: 10.1152/ajplung.00067.2004. [DOI] [PubMed] [Google Scholar]
- Hickerson R P, Smith F J, Reeves R E, Contag C H, Leake D, Leachman S A, Milstone L M, McLean W H, Kaspar R L. Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J Invest Dermatol. 2008;128:594–605. doi: 10.1038/sj.jid.5701060. [DOI] [PubMed] [Google Scholar]
- Nakai N, Kishida T, Shin-Ya M, Imanishi J, Ueda Y, Kishimoto S, Mazda O. Therapeutic RNA interference of malignant melanoma by electrotransfer of small interfering RNA targeting Mitf. Gene Ther. 2007;14:357–365. doi: 10.1038/sj.gt.3302868. [DOI] [PubMed] [Google Scholar]
- Gaither L A. Chemogenomics approaches to novel target discovery. Expert Rev Proteomics. 2007;4:411–419. doi: 10.1586/14789450.4.3.411. [DOI] [PubMed] [Google Scholar]
- Kawasumi M, Nghiem P. Chemical genetics: elucidating biological systems with small-molecule compounds. J Invest Dermatol. 2007;127:1577–1584. doi: 10.1038/sj.jid.5700853. [DOI] [PubMed] [Google Scholar]
- Verheijen F W, Verbeek E, Aula N, Beerens C E, Havelaar A C, Joosse M, Peltonen L, Aula P, Galjaard H, van der Spek P J, Mancini G M. A new gene, encoding an anion transporter, is mutated in sialic acid storage diseases. Nat Genet. 1999;23:462–465. doi: 10.1038/70585. [DOI] [PubMed] [Google Scholar]
- Helip-Wooley A, Kleta R, Gahl W A. Lysosomal free sialic acid storage disorders: Salla disease and ISSD. Barranger J A, Cabrera M, editors. New York, NY, USA: Springer; Lysosomal Storage Diseases. 2007:499–511. [Google Scholar]