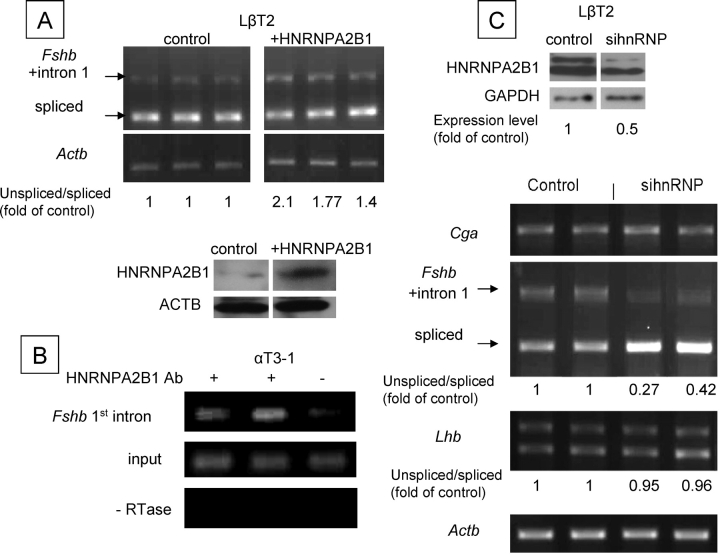
FIG. 6.
Ribonucleoprotein A2/B1 represses Fshb first intron splicing. A) After overexpression of HNRNPA2B1 in LβT2 cells, RT-PCR was carried out to evaluate the spliced and unspliced Fshb mRNA using primers on the first and second exons. The relative levels of unspliced:spliced transcripts were measured by densitometry readings, and are shown as fold difference of the level in control cells. The level of HNRNPA2B1 overexpression, assessed by western analysis, is also shown. B) RIP was carried out in αT3–1 cells using antisera to HNRNPA2B1 and primers that span the first intron of Fshb. C) SiRNA constructs targeting Hnrnpa2b1 were transfected into LβT2 cells, and Western blots confirm the reduced levels of HNRNPA2B1; also shown are the quantified levels relative to those in controls, after normalization to glyceraldehyde phosphate dehydrogenase (a sum of both isoforms). The effects of these constructs on each of the gonadotropin subunits were assessed by RT-PCR using primers that span an intron, and quantified as in A. The band for the Cga transcript is the spliced form (due to the larger intron and the short amplification time used in the PCR).