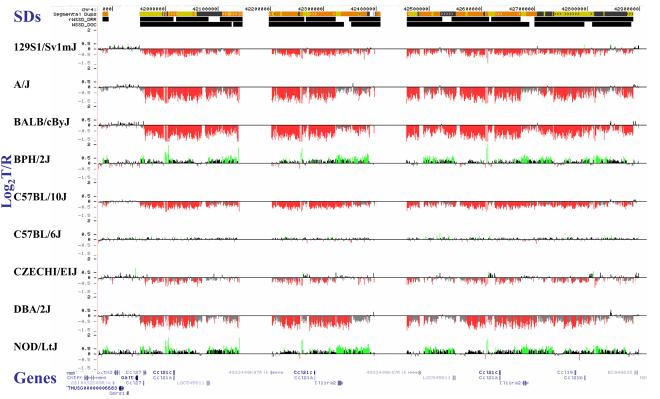
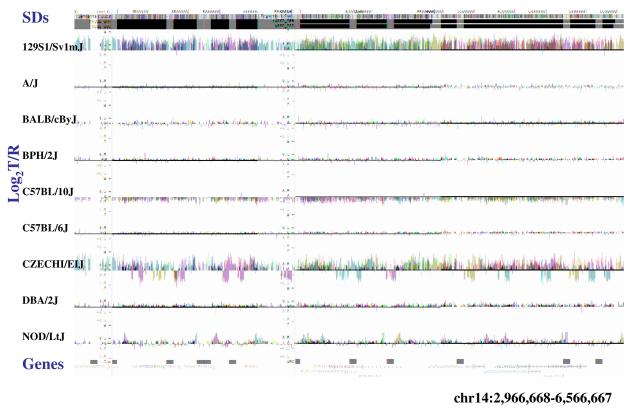
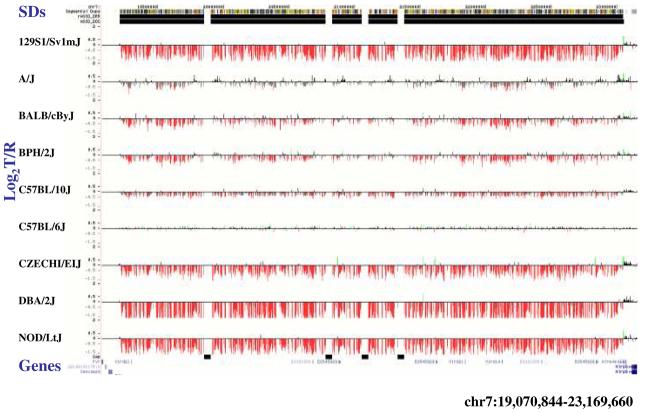
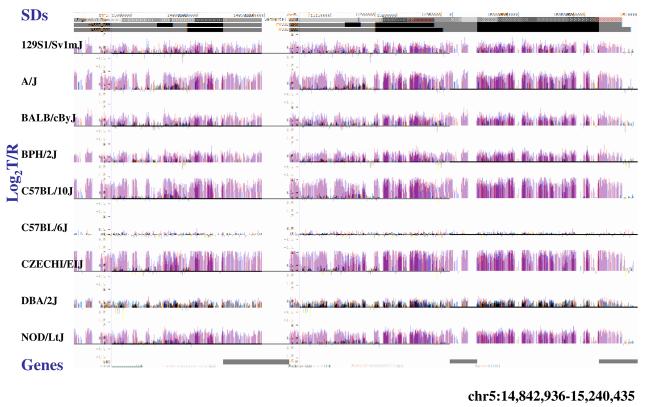
Figure 4. Copy-number variable mouse segmental duplications.
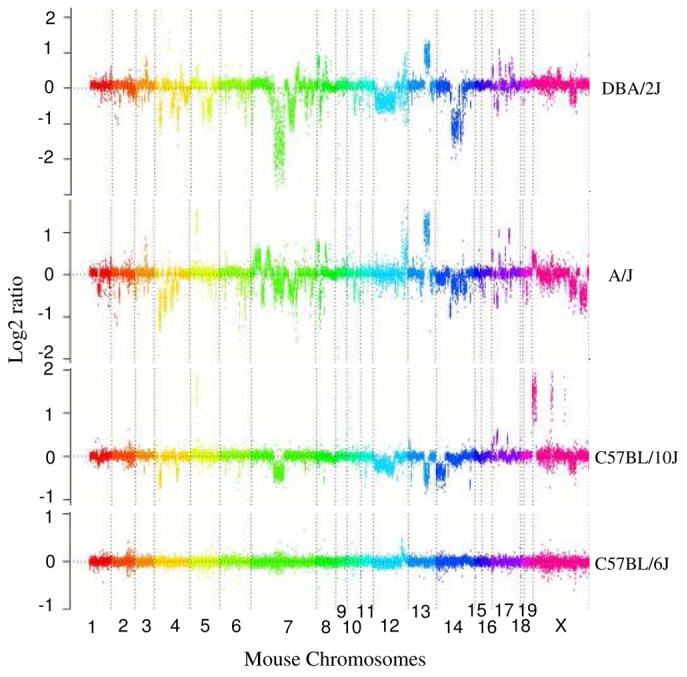
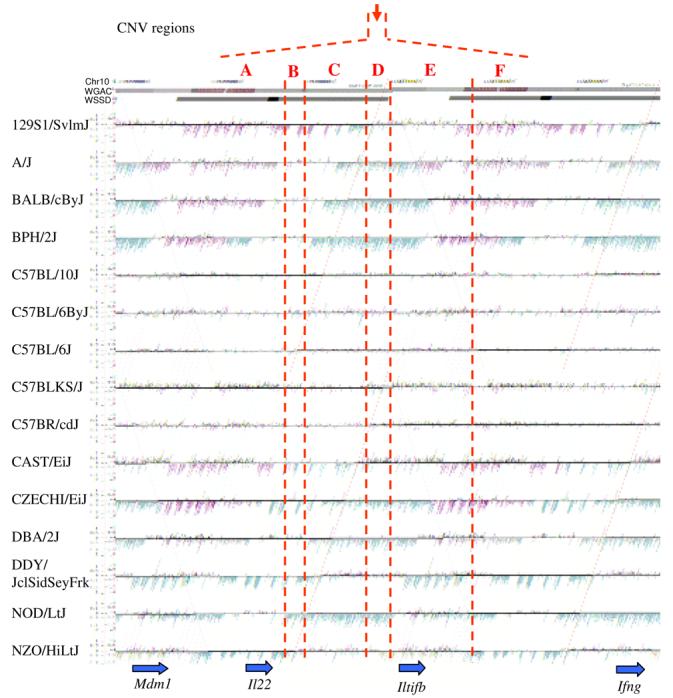
(a) Underlying array comparative genomic hybridization data are shown for four strains compared to C57BL/6J. SD and flanking regions (159 Mb) were ordered and collapsed according to chromosomal position (color). (b) An ∼170 kb segmental duplication region on chromosome 10 shown from the browser (http://mouseparalogy.gs.washington) in more detail for 15 different mouse strains. Significant (>1.5 standard deviation) decreases (red) and increases (green) are highlighted. At least six distinct regions (A-F) of copy-number variation can be discerned within the duplication block (WGAC=whole-genome assembly comparisons, WSSD=whole-genome shotgun sequence detection). Region A & E represent high-identity duplications of the interleukin 22 gene and, therefore, the arrayCGH signal represents the average differential of both regions and the arrayCGH patterns mirror one another. (c-f) Other examples of copy-number variable regions of segmental duplication depicted for nine strains, including: (c) the CCl and Il11ralpha duplication block (chr4:41,687,499-42,962,500), (d) a Spetex duplication block (chr14:2,966,668-6,566,667), (e) a vomeronasal receptor (V1r) duplication block (chr7:19,070,844-23,169,660), and (f) a Speer4d gene family duplicated region (chr5:14,842,936-15,240,435).