Figure 6.
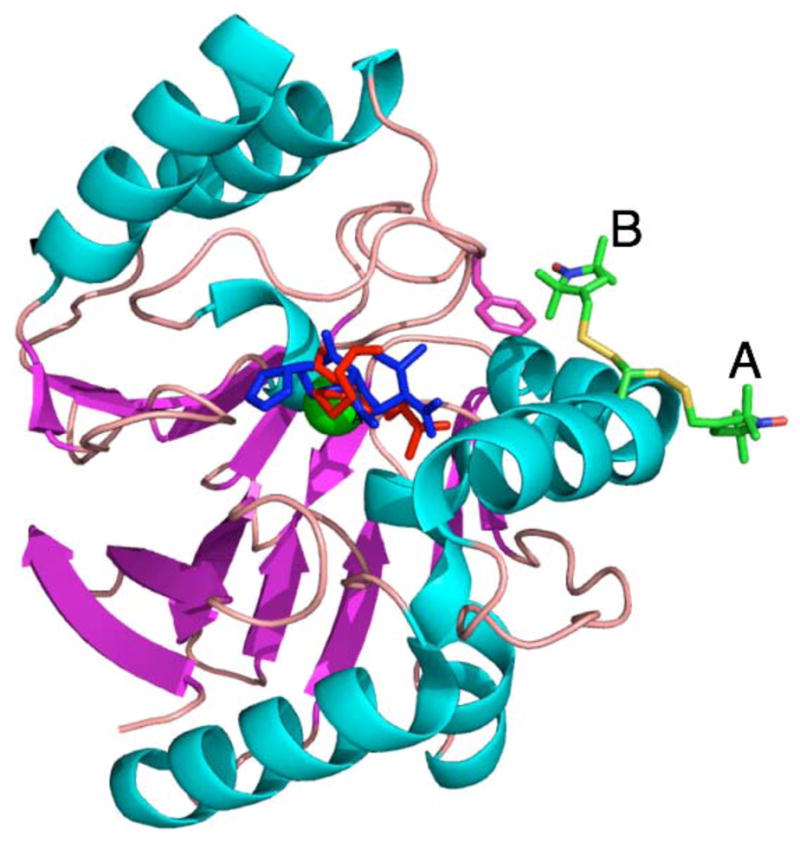
A model for ImiS* based on the structure of the CphA complex with the hydrolysis product of biapenem. A Glu152Cys in silico mutation (MacPyMol v.0.99) was carried out on the CphA structure (PDB-ID 1X8I) and the Cys152 side chain was replaced by a Cys-MTSL fragment from the structure of spin-labeled T4 lysozyme (PDB-ID 1ZYT). Two conformations of Cys-MTSL are shown, A and B, that correspond to rotation around the X1 (Cα-Cβ) dihedral axis. Imipenem (red) was manually modeled onto biapenem (blue) to preserve, as far as possible, both the Zn binding and the presumed orientation of the lactam ring. The Zn(II) ion is shown in green in the center of the figure and the side chain of Phe236 is shown in magenta.
