Fig. 1.
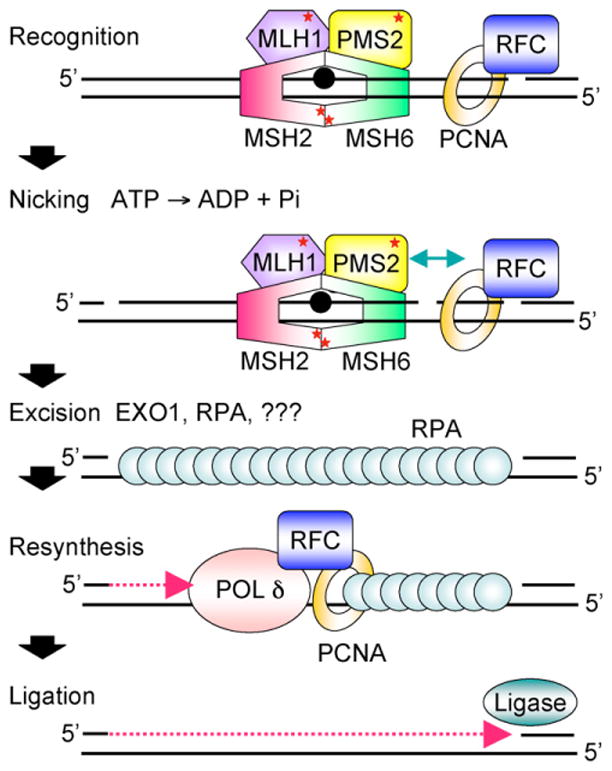
Cartoon scheme for 3′-directed eukaryotic MMR. Recognition of a mismatch by MutSα (MSH2-MSH6) or MutSβ (MSH2-MSH3, not shown) and MutLα (MLH1-PMS2) results in the formation of a ternary complex whose protein-protein and protein-DNA interactions are modulated by ATP/ADP cofactors bound by MutSα and MutLα (indicated by red *). PCNA may play an important role in the recruitment of MMR proteins to the vicinity of the replication fork via a PIP motif on MSH6 and MSH3. Nicking by the endonuclease function of PMS2 stimulated by ATP, PCNA, and RFC and relevant protein-protein interactions (indicated by green arrow) may establish strand discrimination targeting repair to the newly synthesized strand. MMR is bidirectional and can be 5′-directed as well; this is not shown. HMGB1, a nonhistone chromatin protein that bends DNA also facilitates MMR in vitro at or before the excision step (not shown). Excision by EXO1 and possibly other as yet unidentified exonucleases leads to the formation of an RPA-coated single-strand gap. Resynthesis by replicative polδ and ligation restore the integrity of the duplex. See text for details.
