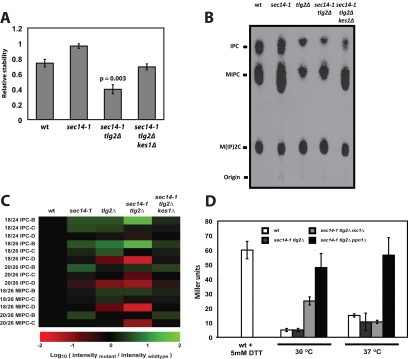
Figure 8.
Ceramide homeostasis and enhanced catabolism of inositol SLs. (A) Inositol-SLs are labile in sec14-1ts tlg2Δ double mutants. Wild-type, sec14-1ts, sec14-1ts tlg2Δ, and sec14-1ts tlg2Δ kes1Δ yeast were radiolabeled to steady state with 3[H]inositol (20 μCi/ml) at 30°C. Subsequently, cells were washed and resuspended in glucose-SD media and incubated for 3 h at 37°C in the presence of unlabeled inositol (12 μM). Lipids were extracted from cells and subject to alkaline methanolysis. Inositol-SLs were quantified by liquid scintillation counting and relative inositol-SL stability for each strain is expressed as the ratio of [3H] after chase:[3H] before chase. Values represent averages (and standard deviations) from three independent experiments. (B) Inositol SL profiles are altered in tlg2Δ mutants. Wild-type, sec14-1ts, tlg2Δ, sec14-1ts tlg2Δ, and sec14-1ts tlg2Δ kes1Δ yeast were labeled to steady state with [3H]inositol (20 μCi/ml). After a 2-h shift to the restrictive temperature for the sec14-1ts allele (37°C), lipids were extracted from 5 OD600 nm of cells and subject to alkaline methanolysis. TLC profiles for the inositol-SLs for yeast strains of indicated genotype are shown and each lipid assignment (indicated at left) was determined by Rf. (C) Quantitative inositol SLomics. Complex SLs were extracted from 25 OD600 of yeast and analyzed by mass spectrometry. Lipid mass was calculated by comparison with internal standards. Each SL species is represented as the log10 of the mutant:wild-type ion intensity ratio. Species shown in red report decreased mass in mutant relative to wild-type, whereas green indicates increase. (D) Incorporation of a ppn1Δ allele into the sec14-1ts tlg2Δ genetic background restores UPR activity. UPR activity was monitored via the UPRE::LACZ reporter in wild-type, sec14-1ts tlg2Δ, sec14-1ts tlg2Δ isc1Δ, and sec14-1ts tlg2Δ ppn1Δ yeast strains as indicated. Yeast were cultured at the permissive temperature (30°C) to mid-logarithmic growth phase and then shifted to the restrictive temperature (37°C) for 2 h before analysis. As control for UPR activity, wild-type yeast were challenged with DTT (final concentration, 5 mM) for 60 min before assay.