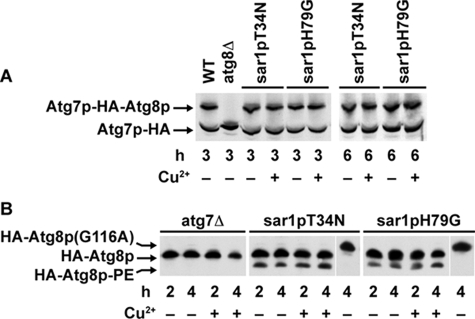
Figure 6.
Effects of Sar1p on the lipidation of Atg8p. (A) atg7pC518S-HA was expressed in GS115, SJCF257 (atg8Δ), WDY64, and WDY65 cells. The cells were adapted from YNM to YND in the absence and presence of CuSO4 for 3 and 6 h. At that time, the cells were solubilized in SDS sample buffer, and the 70-kDa Atg7p and 95-kDa Atg7p protein–protein ester conjugate visualized by Western blotting. The protein–protein conjugates of atg7pC518S were observed in wild-type cells and in cells expressing sar1pT34N or sar1pH79G, but not in cells lacking Atg8p. (B) atg7Δ, WDY64 (sar1pT34N), and WDY65 (sar1pH79G) cells expressing HA-Atg8p were adapted from YNM to YND in the absence and presence of CuSO4 for 2 or 4 h. The cells were then solubilized, proteins separated on 12% SDS-PAGE 6 M urea gels, and the molecular forms of Atg8p were detected by Western blotting using mouse anti-HA antibodies. During 2–4 h of glucose adaptation, two forms of HA-Atg8p were apparent in the sar1pT34N and sar1pH79G cells. The faster migrating form of HA-Atg8p was absent in atg7Δ cells and in cells expressing HA-Atg8p(G116A). HA-Atg8p(G116A) cannot be lipidated because of its inability to be proteolytically processed and thus migrates slower than HA-Atg8p (Mukaiyama et al., 2004). Based on these findings, we have identified this faster migrating band as the lipidated form of Atg8p (HA-Atg8p-PE).