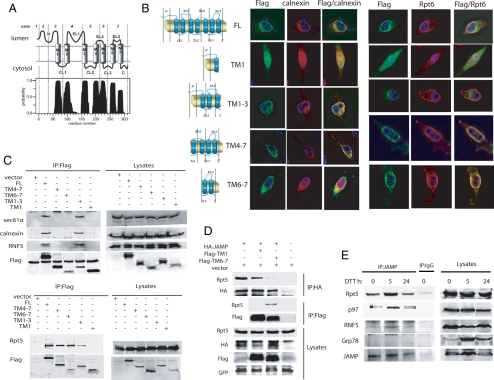
Figure 2.
JAMP is an ER stress–inducible protein that associates with ERAD and proteasome components using N- and C-terminal domains, respectively. (A) TMHMM-based prediction for the membrane organization of JAMP. Possible position of JAMP C- and N-terminal domains within the cytosol and lumen, respectively, is outlined based on the TMHMM program. (B) Colocalization of the C-ter and N-Ter fragments of JAMP with ER and proteasome components. Truncated fragments representing different domains of JAMP (schematic outline on the left) were generated based on structural prediction and assessed for colocalization with endogenous calnexin or Rpt6. Flag-tagged JAMP domains were transfected into HeLa cells, and immunostaining was performed 24 h later with the indicated antibodies. (C) The C-terminal of JAMP binds proteasome subunits, whereas the N-terminal binds ERAD components. Deletion mutants of JAMP (Flag-tagged) were transfected into 293T cells, and 24 h later proteins were prepared and IPed with Flag antibodies. Immunoblots were carried out using the indicated antibodies against endogenous Rpt5, Sec61α, calnexin, and RNF5. (D) Overexpression of the JAMP C-terminal disrupts interaction between JAMP and proteasome subunits. HA-JAMP was cotransfected with Flag-TM 1, Flag-TM 6–7, or vector as control. After 36 h cells were lysed and subjected to IP with HA antibody after immunoblot analysis with Rpt5 and HA antibodies. Lysates were also subjected to parallel IP with Flag antibodies to compare interaction of Rpt5 with C-terminal (TM 6–7) versus N-terminal (TM1) JAMP. GFP expression served as control for transfection efficiency. (E) ER stress increases endogenous JAMP expression and its association with Rpt5, RNF5, and p97. 293T cells subjected to ER stress (10 mM DTT for 30 min), and proteins were prepared (lysis buffer supplemented with 1% Triton X-100) at the indicated time points. Left, IP with anti-JAMP antibody was followed by immunoblot analyses using indicated antibodies; right, total lysates.