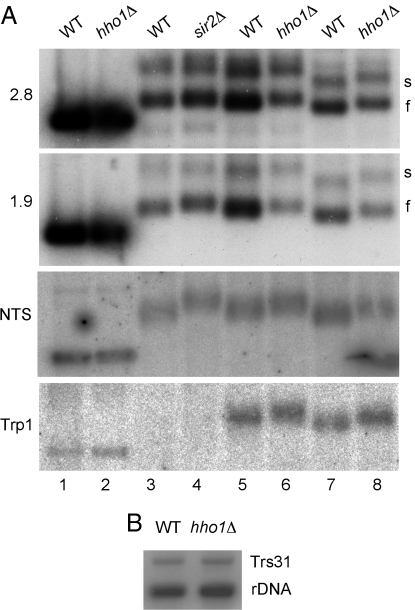
Fig. 3.
Deletion of Hho1 leads to increased chromatin accessibility to psoralen. (A) Psoralen cross-linking of rDNA. Wild-type, hho1Δ, and sir2Δ logarithmic cell cultures were exposed to 10 μg/ml psoralen and 365-nm UV light for 40 min. DNA was extracted, digested with EcoR1, electrophoresed on a 1.3% agarose gel, the cross-linking reversed and blotted onto a nitrocellulose membrane. The blot was probed with probes that recognize 1.9 and 2.8 kb fragments of the 35S rRNA transcript, the rDNA NTS, and the TRP1 gene. Strains used for the SIR2 control were deleted for TRP1. Blots were exposed to a phosphorimager screen. Cultures in lanes 1–6 were harvested at 0.3 OD600, lanes 7 and 8 at 0.9 OD600. (B) Copy number comparison of wild-type and hho1Δ strains. Extracted DNA was digested with EcoR1 and BamH1, electrophoresed, and blotted as above. The blot was probed with a mixture of two probes recognizing the 1.9-kb rDNA fragment and single-copy TRS31 DNA. Quantitation of the bands showed an rDNA/TRS31 ratio of 4.31 in the wild-type and 3.74 in the hho1Δ strain.