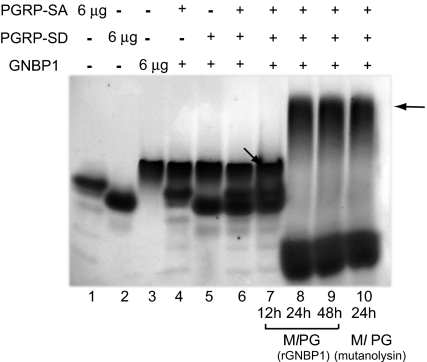
Fig. 4.
Ternary complex formation in the presence of PG. Processed PG shifted all of the three proteins into a higher-molecular-weight complex in native Tris-glycine PAGE gels. 6 μg of rPGRP-SA, rPGRP-SD, and rGNBP1 were used in single protein species controls (lanes 1–3). rGNBP1 mixed with either rPGRP-SA or rPGRP-SD as a 1:2 molar ratio is shown in lanes 4 and 5. The rPGRP-SA, rPGRP-SD, and rGNBP1 mix at 2:2:1 ratio is shown in lane 6. PG from Ml processed by rGNBP1 as described during 12 h (lane 7), 24 h (lane 8), or 48 h (lane 9) was incubated with the 3-protein mix (as defined in lane 6). Protein mix incubated with PG digested completely by mutanolysin (24 h) was used as control (lane 10). Arrows illustrate potential protein complexes. Lower bands in lanes 8, 9, and 10 represent PG fragments bound to PGRP-SA. In this form PGRP-SA will be running faster than the loading control of protein alone because AUC data indicate that it becomes more spherical upon microbial ligand addition (see Table S1, where S value increases from 2.0 to 2.2 upon muropeptide addition). Protein bands were visualized by Coomassie blue staining.