Fig. 3.
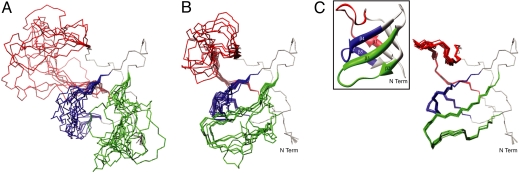
Solution structure of the invisible, Ark1p-peptide bound conformation of the Abp1p SH3 domain. (A) Ensemble of 10 starting structures generated from high temperature molecular dynamics of the apo-Abp1p SH3 domain x-ray structure (22), as described in the text. Regions in gray are fixed to the x-ray structure, because ΔϖRMS ≈ 0. The pair-wise rmsd values of the backbone Cα, CO and N atoms of regions 1 (red), 2 (blue) and 3 (green) are 5.8 ± 1.6, 3.7 ± 1.0, 2.6 ± 0.7 Å, respectively (10.4 ± 1.6, 8.6 ± 1.2, 3.8 ± 0.6 Å with respect to the apo-Abp1p1 SH3 domain x-ray coordinates). (B) Ensemble of the 10 lowest energy structures generated using (φ, ψ) restraints exclusively, as described in the text. Pair-wise rmsd values of regions 1, 2 and 3 are 3.1 ± 1.4, 3.8 ± 2.4 and 1.2 ± 0.7 Å, respectively. (C) As in B, but including restraints from residual anisotropic interactions as measured using a single alignment media (Pf1). The rmsd values of 0.39 ± 0.12, 0.30 ± 0.11 and 0.20 ± 0.06 Å are calculated for regions 1–3 (0.47 ± 0.10, 0.76 ± 0.14 and 0.53 ± 0.07 Å to the reference structure of the bound form). Inset, ribbon diagram of the apo-Abp1p SH3 domain x-ray structure; all conformers in the figure are in the same orientation. All of the rmsd values reported are calculated by superimposing the “fixed” regions (gray in Fig. 2C); the fixed regions are not included in the computation.