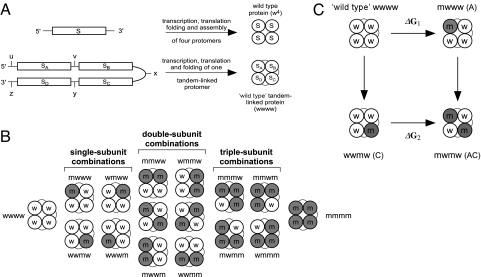
Fig. 1.
Strategy for a direct analysis of cooperativity in multisubunit allosteric proteins. (A) Schematic representations of monomeric and tandem tetrameric gene constructs giving rise to assembled tetrameric protein particles, one in which the subunits are not linked to each other and another where the identical subunits are tandem-linked. The identical subunits of the tandem tetrameric gene are separated by unique restriction sites, as indicated by the lowercase letters. (B) Such a gene design allows the construction of all possible combinations of mutated subunits. Lowercase letters w and m designate wild-type and mutated subunits, respectively. (C) Thermodynamic double-mutant cycle analysis applied to calculate the magnitude of intersubunit coupling free energy between subunit pairs. According to this analysis, the equilibrium effect on protein function upon mutating one protein subunit is evaluated once, when the other subunit is native (ΔG1) and again when it has been mutated (ΔG2). If the two subunits operate independently, then the conformational change of one subunit is independent of the conformational state of the adjacent subunit and Δ2Gi,j (= ΔG1 − ΔG2) equals zero. Otherwise, the two subunits under question can be considered as being coupled, and Δ2Gi,j ≠ 0.