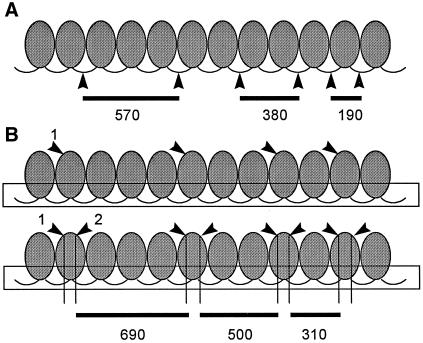
Figure 8.
A model to explain the unusual multimeric sizes of the Tec element chromatin MNase digestion products. (A) The normal pattern of digestion of a nucleosomal array by MNase along with fragment sizes predicted for nucleosomes that show a repeat spacing of 190 bp. (B) An alternative pattern of cutting that could give rise to the unusual multimers seen for the Tec elements. Instead of preferential digestion in the spacer regions, we propose a pattern of cutting within the nucleosomal core–associated DNA. We assume that a higher-order chromatin structure (shown as a box surrounding the spacer regions) prevents accessibility of MNase in the spacer region. We propose that asymmetry in the cut sites that give rise to the multimers arises because multiple cuts occur within a core, leaving behind portions of nucleosomal core–associated DNA. This could happen in a stepwise manner as shown, in which one site within the core is preferred and then additional sites within the core become sensitive after the first site is attacked. If the amount of DNA removed by cutting within the core-associated DNA is approximately one loop, or 78–79 bp, the multimers would be ∼110 bp larger than those produced from the pattern shown in A. Note that both patterns of cutting give rise to multimers that differ by 190 bp, as seen for both the Tec element and mac-destined sequences.