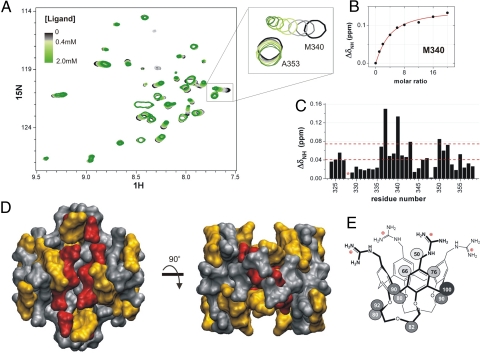
Fig. 5.
Protein p53TD mapping. (A) 1H-15N-HSQC overlaid spectra for the titration of p53TD with ligand 1. Only the central region is shown. Color code: in black, the free protein (125 μM tetramer); in the gray scale, up to 500 μM ligand; in the green scale, from 500 μM to 2.0 mM ligand. (B) Perturbation of Met-340 resonance adjusted to a simplified 1:1 binding model (red line) (see Methods for details). (C) Chemical shift mapping at the end of the titration (*, not available data). Dashed red lines drawn across indicate the cutoff limits used to categorize the perturbation degrees for mapping the 3D protein structure represented in D: in red, residues shifted >0.075 ppm (the mean shift plus one standard deviation); in orange, those shifted >0.04 ppm (the mean shift). (E) STD epitope mapping of ligand 1. Saturation normalized percentages are displayed for each proton of the molecule; 100% corresponds to the 1H with the highest saturation degree, thus, the one closer to the protein.