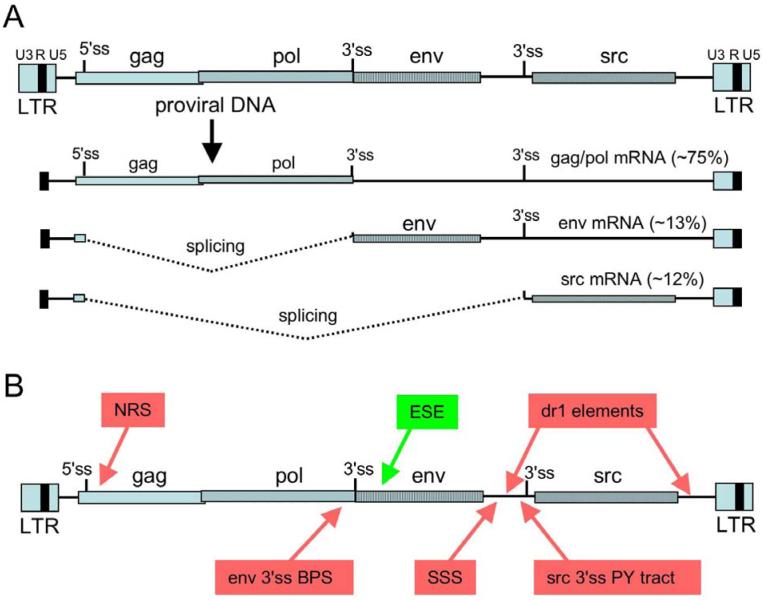
Figure 1.
Elements that influence RSV RNA processing. A) Structure of the RSV provirus and mRNA species. Shown are the LTRs at the termini of the provirus with the U3, R (black box), and U5 regions indicated. The gag, pol, env, and src gene coding regions are shown with different patterned boxes, and the 5′ and 3′ splice sites are indicated. Transcription starts at the beginning of the 5′ R region, and RNAs are cleaved and polyadenylated at the end of the 3′ R region. The relative percentages of unspliced, env, and src mRNA are indicated, and splicing is depicted by dotted lines. B) Summary of cis elements that control splicing in RSV. The schematic of the proviral DNA is as in Figure 1. Red boxes indicate negative-acting elements and include: the negative regulator of splicing, or NRS; the suboptimal BPS associated with the env 3′ ss; the suppressor of src splicing (SSS) that specifically represses src splicing; dr1, the direct repeat elements that flanks the src gene and specifically repress src splicing; the suboptimal pyrimidine tract associated with the src 3′ ss. Splicing to env is also controlled by a positive-acting exonic splicing enhancer (green). Adapted and updated from (20).