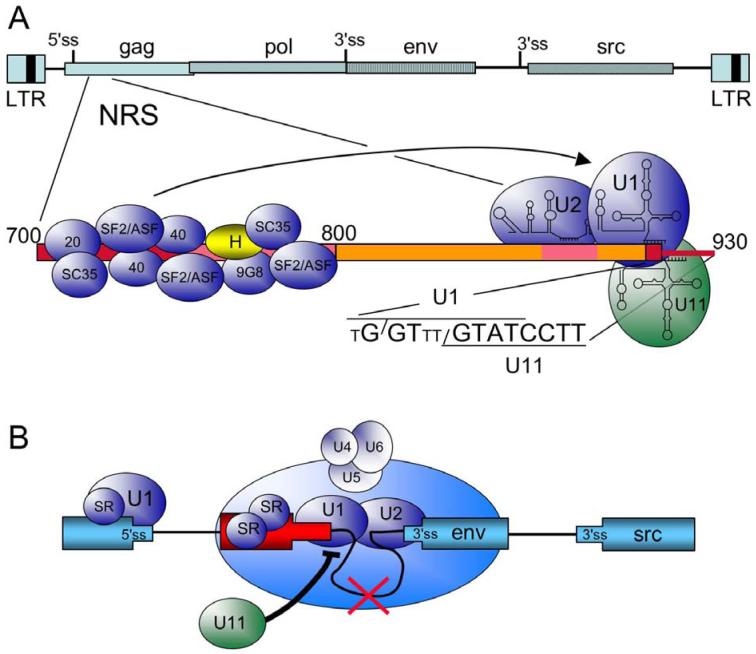
Figure 2.
Summary of cis elements and trans factors important for NRS splicing control. A) At top is a schematic of the RSV proviral DNA (as in Figure 1) and below is an expansion of the NRS (nt 700 to 930). Regions important for NRS splicing inhibition are boxed in red and pink. NRS5′, from nt 700 to 800, binds the indicated SR proteins (blue) and hnRNP H (yellow). U1, U2, and U11 snRNPs bind to the distal end of NRS3′ (nts 800 to 930). The sequence of the degenerate U1 binding site (small T represents non-consensus residues) and the perfect U11 binding site are shown. Slashes indicate the splice site for each sequence. U2 snRNP most likely binds to an upstream BPS-like sequence (not shown). The arrow indicates that the SR protein promote U1 binding to NRS3′. B) Model for NRS splicing inhibition. In the schematic, the RSV exons are in blue, and the NRS is in red. SR proteins promote U1 binding to the authentic 5′ ss and the NRS, setting up a competition for interactions with the 3′ splice sites (env in this example). As detailed in the text, NRS-bound U1 interacts with the 3′ ss to form an early splicing complex, which then matures into a non-catalytic spliceosome-like complex upon the addition of the U4/U6.U5 tri-snRNP, which is not correctly integrated into the complex. Splicing from the NRS fails (red X), and the complex sequesters the 3′ ss from a productive interaction with the authentic 5′ ss. U11 binding would block U1 binding to the overlapping site (thick black line), preventing assembly of the NRS-3′ss complex, and allowing normal splicing to occur.