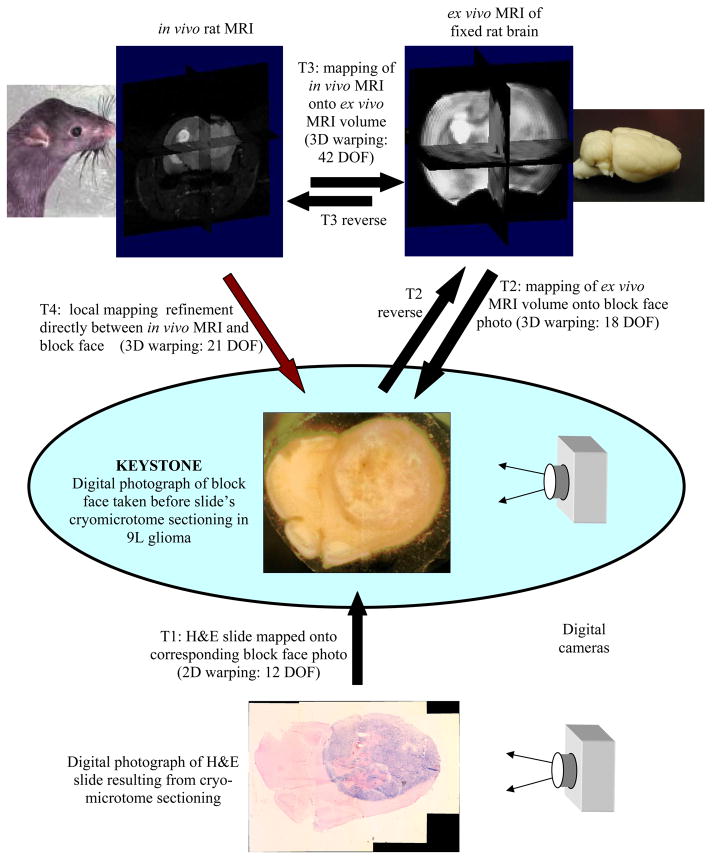
Abstract
We present a method for registering histology and in vivo imaging that requires minimal microtoming and is automatic following the user’s initialization. In this demonstration we register only one hematoxylin and eosin (H&E) stained histological slide containing a 9L glioma brain tumor in a rat with a diffusion weighted, in vivo 7T MRI. Since the spatial resolution of the in vivo diffusion-weighted MRI is limited, we add the step of obtaining a high spatial resolution, ex vivo MRI in situ for intermediate registration. In general we follow the lead of Kim [1] in the application of maximizing mutual information to optimize the registration between all pairings of image data whether the sources are MRI, tissue block photograph, or stained sample photograph. The warping interpolant used is thin plate splines [2] with the appropriate basis function for either 2D or 3D applications. All registrations are implemented by user initialization of the approximate pose between the two data sets, followed by automatic optimization based on maximizing mutual information. While only the Lo-b or Hi-b components of the in vivo MRI volume are used in the registration process, the calculated ADCo values from the registered location appear highly correlated with the H&E slide.
Keywords: Registration, mutual information, warping, block face, histology, slide, volumetric MRI, thin plate spline, optimize
Introduction
The goal of correlating in vivo imaging results with histology has been sought for some time via methodology that continuously evolves. This generic problem has at least two major components that must be addressed by all methods: 1) correcting for the 2D distortions in the preparation and cutting of the histology specimen from the tissue block, and 2) restoring the 3D geometry of the tissue block back to its in vivo state. Typically both steps require complex geometric transformations characterized by warping. A relatively quick survey of such literature over the last 10 years yields a variety of methods. In 1997 Kim [1] demonstrated an automatic method for warping neuroautoradiographic slices back to their geometry in the tissue block and then one final manually initialized 3D warping from the geometry of the tissue block to its geometry in the in vivo brain MRI image using mutual information [3, 4] as the objective function and thin plate splines as the geometric warping interpolants [2]. In 1999 Jacobs [5] described a sequential technique first using Pelizzari’s automatic surface fitting method for rigid structures [6] followed by manually defined warping using thin plate splines. In 2003–2004 Wilson’s group has published several papers describing the use of manually inserted needle tracks and the iterative closest point method [7] to register in vivo MRI with tissue slices in 3D followed by the use of manually chosen homologous points augmented with points on contours to warp the 2D histology slices back into the geometry of the tissue slices [8–10]. In 2004 Zarow describes postmortem MRI and brain tissue registration obtained using a technique using the Pearson cross-correlation coefficient first and mutual information last as two separately and sequentially applied objective functions and an nth order polynomial as a 2D (only) geometric warping interpolant[11]. In the following paragraphs we describe an automated method that is relatively free from user bias in locating homologous points in that it is driven by maximization of mutual information, can operate on multimodal data, and can handle mixed combinations of 2D and 3D datasets.
Methods
In the following we describe how we can map the location of a highly magnified histology section whose total field of view is typically smaller than an MRI voxel back to a computed ADCo voxel location in the in vivo MRI without the need for manual creation in tissue or definition of homologous points in images based on intensity variations; manually placed control (not homologous) points are used only to initialize a mutual information driven optimization. Here we emphasize the difference between the function of “homologous” and “control” points. By definition homologous points represent identical morphological structures within multiple volumes chosen either by an algorithm or human; on the other hand control points in splines are simply “handles” which the optimizer can move as hypothesized homologous points, while testing the hypothesis by searching for the optimal solution as defined by an objective function, i.e. mutual information in our case.
We first present a schematic overview that describes in broad terms the scope and rationale behind the technique. Following the schema presentation we add detailed information regarding each of the labeled steps in the schema, and thus present the full story of how a microscopic region of a histology slice can be mapped back to the single voxel in which it resided during in vivo imaging of the animal. In the following text we will interchangeably use the phrases “mapped onto” and “registered with”, e.g. the “floating or homologous” data set is “mapped onto or registered with” the reference dataset.
The preceding overview schema is a summary of how a histological slide can be mapped back to its location in an in vivo MRI volume. In the following paragraphs we will add detail to the methodology of the individual steps. Note that the keystone in this whole process is the tissue block face image acquired before the tissue slice is cut by the microtome, as it will ultimately drive all of the important registrations which we describe. The block face and H&E slide images were acquired by digital photography; the contrast in the block face image was increased using histogram equalization. The in vivo MRI was a 128×128×13 slice, diffusion, spin-echo, multislice scan, TR = 3s, TE = 60 ms, fov = 3×3 cm2 of 1 mm slices with 0.5 mm gap. The ex vivo MRI is a high resolution 3D FSE, 128×128×32 slices, echo train = 16, TR = 4 s, TE = 15 ms, and fov = 2×2×2 cm3 acquired by imaging the in vitro fixed specimen suspended in normal saline over several hours to obtain good signal to noise ratio.
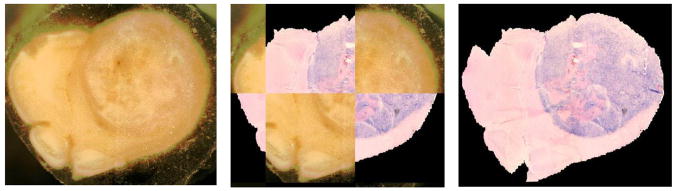
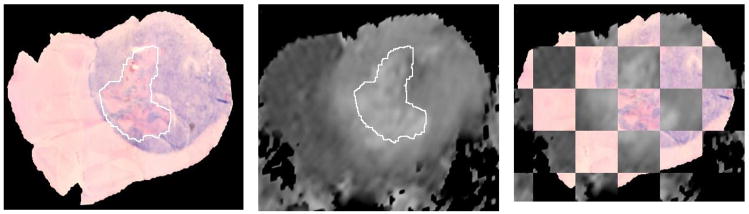
We begin at the bottom of the Overview Schema, Fig. 1. After the histological slide is cut from the tissue block it is inevitably deformed in a manner relatively uncorrelated with other adjacent cuts only tens of microns away. When the slide is not torn, this deformation can be recovered, i.e. unwarped, by registering the resulting (H&E in this example) slide back onto the geometry of the block face. This transformation is reliably performed as a 2 dimensional (2D) thin plate spline (TPS) warping using 5 – 10 control points (here we use 7) using mutual information as the objective function [1]. In [1] as well as here the control points are initially approximately uniformly distributed on the reference block face image while only the first 3 are placed by the user in approximate homology on the floating image for initialization. While the reference control points remain stationary, the optimizer drives the loci of the control points in the floating image, i.e. the H&E slide in this case, first using only 3 control points to approximate the transform as a (linear) full affine transform, and then finally using all 7 to warp the floating image such that the mutual information between the two datasets is maximized [12]. Fig. 2 shows the mapping accuracy achieved in restoring the original H&E slide shown in the Schema above to the geometry of the block face. In addition to showing the geometrically restored H&E stained tissue sample in Fig. 2C, we have used a common “checkerboard” display in Fig. 2B to show the alignment of multiple structures where each alternate checkerboard block comes from the alternate image. The computed transformation obtained by the optimal registration is denoted as T1 in the Overview Schema, Fig. 1.
Fig. 1.
Overview Schema for Registration of Histology with in vivo MRI
Fig. 2.
Fig. 2A. Picture of tissue block face in cryomicrotome before cutting the slide sample.
Fig. 2B. Slide resulting from cut, H&E stained, and mapped onto block face image (2D T1 transform, 12 DOF)
Fig. 2C. H&E slide restored to block face geometry as a result of the T1 transformation
Now that the geometry of the histology specimen has been restored to that of the tissue block we will concentrate on ultimately mapping the correct 2D manifold within the 3D in vivo MRI onto the block face image (and consequently onto the H&E slide). Due to the lower spatial resolution of the in vivo MRI and the dramatic change in geometry after the brain was been removed from the cranium, we have elected to register the block face with an intermediate ex vivo MRI scan rather than directly proceeding to registration with the in vivo MRI. In a sense this replaces the complete serial sectioning of the block face to obtain the ex vivo geometry as we reported in our earlier publication [1]. The intermediate anatomical MRI scan is acquired from the ex vivo tissue specimen after it has been removed from the cranium before mounting on the cryomicrotome’s cutting block. In vivo the 9L tumor exerts significant internal pressure on the brain, was seeded relatively close to the surface in the brain, and is essentially non-invasive. The result is that when the brain is removed from the cranium the tumor behaves a little like popcorn in that it literally explodes in size while the remainder of the brain decompresses as well. For an example of the dramatic change see Fig. 5A and recall that the dotted line approximates the surface of the brain as constrained by the cranium in vivo.
Fig. 5.
Fig. 5A. Anterior axial slice of ex vivo reference volume. (Dotted line approximates the restraining boundary of the cranium in vivo.)
Fig. 5B. Checkerboard showing mapping of in vivo MRI onto the ex vivo Lo-b MRI
Fig. 5C. View of in vivo Lob MRI mapped onto the ex vivo MRI.
Fig. 5D. Color overlay of the ex vivo MRI with the grayscale Lo-b in vivo MRI is shown as another way of viewing registration quality.
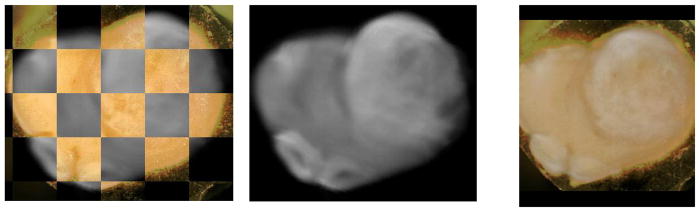
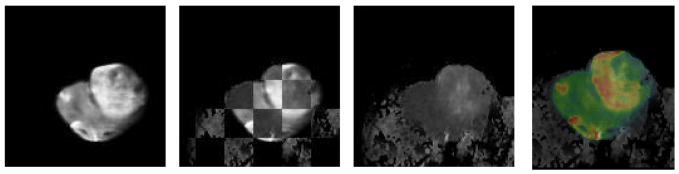
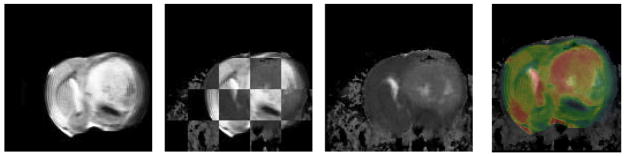
In Fig. 3 we demonstrate the results of mapping a 2D manifold in the 3D ex vivo MRI volume onto block face image using a 6 control point, 3D TPS warping automatically optimized using mutual information as the objective function [12]. This is computed by creating a thin 3D “sandwich” from the 2D block face image and its replica, and warping the 3D in vitro MRI volume such that its intersection with the 3D sandwich maximizes mutual information. The result of the mapping transformation, identified as T2 in the Overall Schema, of the ex vivo MRI is shown in Fig. 3B and the quality of the registration is shown both in Figs. 3A & C. Additionally in Figs. 4A–E we show the resulting reverse mapping of a finite thickness of the block face image back onto the individual adjacent slices of the ex vivo MRI volume. In Fig. 4 we observe that the manifold is indeed warped in a regular fashion consistent with a smoothly deformed 2D manifold.
Fig. 3.
ex vivo MRI mapped onto block face via T2 transform (3D, 18 DOF)
Fig. 3A. Checkerboard showing registration of ex vivo MRI onto block face image in Fig 1a.
Fig. 3B. Resulting ex vivo MRI mapped onto block face geometry
Fig. 3C. Same as Fig. 3A but registration is shown with overlay of block face in color and ex vivo MRI in gray scale
Fig. 4.
Figs. 4A–E. Show the result of the T2 reverse transform, i.e. block face slice in color mapped onto adjacent ex vivo MRI slices in grayscale.
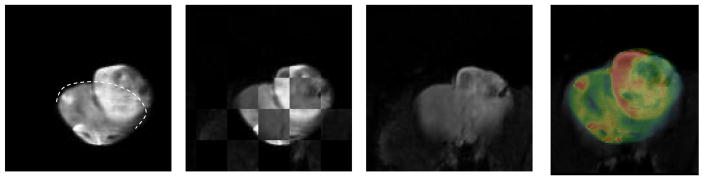
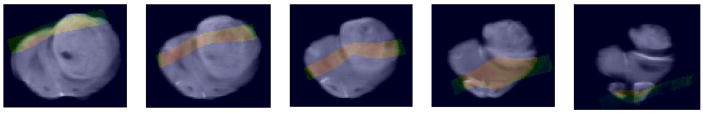
Next we address determining the mapping labeled T3 in the Overview Schema which maps the in vivo MRI onto the geometry of the ex vivo MRI. As mentioned earlier the challenge is to capture the dramatic warping required to map the in vivo MRI onto the dilated lesion in the ex vivo MRI. While the in vivo diffusion MRI is lower resolution, nevertheless the region of support to obtain the registration is sufficiently large and provides enough information for this information-driven technique to succeed as demonstrated in Figs. 5, 6 and 7. Figs. 5 & 6 show the results of the 3D warping computed in the anterior portion of the rat brain which in this case is the region most affected by the tumor size change upon extraction from the cranium in spite of ex vivo, in situ fixation. Fig. 7 demonstrates the same 3D warping that also affects the midbrain section which still contains significant tumor, but is less affected by the dilation. The mapping between the in and ex vivo MRIs was computed using ex vivo MRI as the reference volume and the Lo-b vector component of the diffusion in vivo MRI as the floating volume, and the resulting computed deformation was applied to all three vector components, Lo-b, Hi-b, and ADCo. Figs. 5A–D show the results of mapping the Lo-b diffusion in vivo MRI scan onto the ex vivo MRI scan. Note the high quality of the automatic registration in this anterior region obtained using a 14 control point TPS warping computed by optimizing mutual information between the two data sets (in 3D the degrees of freedom, i.e. DOF = 3n where n is the number of control points). Figs. 6A–D demonstrate the same warping applied to the ADCo vector image component of the in vivo diffusion scan. Figs. 7A–D demonstrate the same 3D warping affecting the midbrain region of the brain. Normally the deformation between two 3D volumes is not as great as in this case, and we are able to fewer control points and use a schedule whose complexity begins only with affine, i.e. linear, transforms and ends with warping, but in this case the deformation is sufficiently great that we must both use more control points and initiate the registration using a manual placement of all 14 control points in the floating data set. The density of control points is highest in the region of the dilated tumor in order to accommodate the needed local warping control.
Fig. 6.
Fig. 6A. Same slice of ex vivo reference volume as shown above
Fig. 6B. Checkerboard showing mapping of in vivo MRI onto the ex vivo ADCo MRI
Fig. 6C. View of in vivo ADCo MRI mapped onto the ex vivo MRI.
Fig. 6D. Color overlay of the ex vivo MRI with the grayscale ADCo in vivo MRI
Fig. 7.
Fig. 7A. Midbrain axial slice of ex vivo reference volume
Fig. 7B. Checkerboard showing mapping of in vivo MRI onto the ex vivo ADCo MRI computed using the Lo-b component
Fig. 7C. View of in vivo ADCo MRI mapped onto the ex vivo MRI.
Fig. 7D. Color overlay of the ex vivo MRI with the grayscale ADCo in vivo MRI
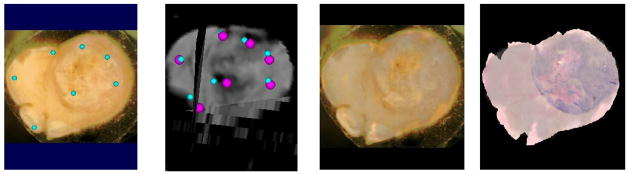
After computing T3 we are nearing our goal of completely defining the mapping from histology to in vivo MRI if we have made no errors in the previous steps. But error-free registration is unlikely across multiple warping transformations, especially where we are dealing with relatively large changes in local deformations and an in vivo MRI of relatively low resolution. Thus we will use what we have estimated thus far to initialize one final registration to improve the resulting geometric accuracy of the final solution. In this last maneuver we will use the reverse of the now estimated T2(T3(in vivo MRI)) transformation to map 7 control points from the reference block face image shown in Fig. 8A back onto a 2D manifold in the 3D in vivo MRI volume as shown by the smaller blue spheres in Fig. 8B. The in vivo MRI Fig. 8B is used as the “floating” image in this registration where the 7 pairs of blue spheres provide the initial estimates of the warping to be computed. Again, a registration of the 3D in vivo data as mapped onto the block face image is computed by optimizing the positions of the control points in the floating image where the objective function to be optimized is the mutual information between the green color component of the block face image and the Hi-b vector component of the diffusion in vivo MRI volume. The Hi-b vector component was chosen for this registration instead of the Lo-b component used in the previous registration due to the increased dynamic range of mutual information between those two components. The pink spheres in Fig. 8B demonstrate the final positions obtained by the optimization of mutual information. The registration obtained is demonstrated by the color overlay shown in Fig. 8C. Note the excellent agreement visualized in Fig. 8C. Fig. 8D is obtained by replacing the block face image in Fig. 8C with the registered H&E slide. The transformation computed in this last step is labeled as T4 in the Overview Schema, and represents the last step required to map any pixel in the H&E slide into a voxel the in vivo diffusion MRI scan.
Fig. 8.
Fig. 8A. Block face reference image with 7 control points; reference points don’t move! (21 DOF mapping)
Fig. 8B. Small blue spheres represent initial mapping of points shown on block face shown on the left into the in vivo Hi-b MRI volume. After one final registration optimization they have been moved to the pink positions
Fig. 8C. Color overlay of block face image with registered grayscale Hi-b in vivo MRI resulting from final control point positions shown in pink in Fig. 8b.
Fig. 8D. Same as 8C, left, except that color block face image has been replaced with the H&E slide mapped onto the block face’s geometry (note the excellent quality of the registration)
Results
Figs. 9A–C emphasize the resulting solution by showing the geometric agreement between the ADCo in vivo MRI scan mapped onto the block face geometry and the H&E slide also mapped onto the block face geometry as shown in Figs. 2B & 8D. In addition to providing a checkerboard image in Fig. 9C to compare edges of structures, we have drawn the identical region of interest (ROI) on Figs. 9A–B. The red hue region in the H&E image represents a region of hemolysis which we would expect to cause increased ADCo values in the corresponding region in the diffusion MRI; that appears to be what we are seeing in this example.
Fig. 9.
Fig. 9. Final Summary figure showing histology slide, i.e. H&E restored to block face geometry, registered to a slice of the ADCo in vivo MRI animal volume. The outlined regions represent identical voxels in each image.
Fig. 9A. H&E slide restored (mapped) to block face geometry. Hemolytic (red) region has been outlined.
Fig. 9B. Same mapping as Fig. 8c–d above is applied to the in vivo ADCo MRI volume. Hemolysis within ROI is consistent with increased ADCo values.
Fig. 9C. Checkerboard verification of registration of in vivo ADCo MRI volume onto H&E slide also restored to block face geometry
Now to summarize, a pixel on the H&E (or any other stained) slide can be marked as the site of a microscopic histology exam/result. That pixel location can be directly mapped onto the block face image via T1, and then into the in vivo diffusion MRI volume using the reverse T4 transformation. Since the TPS solutions possesses no closed form inverse, the reverse, i.e. reversing the role of the data sets and associated control points as reference and homologous images, can be used as the starting point in an iteratively computed inverse using a numerical Newton-based iterative technique.
In summary we have demonstrated a method by which any loci on the histology slide can be mapped back to the corresponding in vivo MRI voxels without significant microtome-hours or compute times (each of the component registrations were obtained in less than 5 minutes except for T3 which required approximately 10 minutes to compute).
There are several possible modifications that we envision which we should explore because they can further increase the robustness of the technique without significantly increasing man-hours or computational burden to perform this registration. For example by acquiring a high resolution in vivo anatomical MRI in addition to the diffusion MRI it may be possible to map directly from the block face image onto the anatomical MRI and eliminate the need for the high resolution ex vivo MRI; the transformation between the two in vivo scans, i.e. high resolution anatomical and the diffusion scan, is essentially a rigid body transform and easily computed. Additionally the use of deeper seeded, more invasive tumors should lead to the need for less dramatic warping between the in and ex vivo conditions. Of course since we are using mutual information as an objective function, in the true theoretic sense the success of this methodology assumes that the tissue being imaged is intrinsically feature rich in all of the modalities; this condition is easily satisfied for most histological studies.
Acknowledgments
This work was funded by DHHS, PHS, NIH, NCI grant 1P01CA85878. The authors would like to thank Tim Desmond and Kirk Frey, MD PhD, for providing the equipment and assistance in cryomicrotome sectionoing and staining of the tissue samples.
Abbreviations
- TPS
thin plate spline
- ADC
apparent diffusion coefficient
- Lo-b
Hi-b, low and high diffusion acquisition gradient acquisitions, respectively
References
- 1.Kim B, Boes JL, Frey KA, Meyer CR. Mutual information for automated unwarping of rat brain autoradiographs. NeuroImage. 1997;5(1):31–40. doi: 10.1006/nimg.1996.0251. [DOI] [PubMed] [Google Scholar]
- 2.Bookstein FL. Principal Warps: Thin-plate splines and the decomposition of deformations. IEEE Trans Pattern Analysis and Machine Intelligence. 1989;11(6):567–585. [Google Scholar]
- 3.Collignon A, Vandermeulen D, Suetens P, Marchal G. 3D multimodality medical image registration using feature space clustering. Proc. CVRMed, Nice, FR. Lecture Notes in Computer Sci. 1995;905:195–204. [Google Scholar]
- 4.Viola P, Wells WM. Alignment by maximization of mutual information. Proc. 5th Int’l. Conf. on Computer Vision, MIT; IEEE Press. 1995. [Google Scholar]
- 5.Jacobs M, Windham JP, Peck DJ, Knight RA. Registration and warping of magnetic resonance images to histological sections. Medical Physics. 1999;26(8):1568–1578. doi: 10.1118/1.598671. [DOI] [PubMed] [Google Scholar]
- 6.Pelizzari CA, Chen GTY, Spelbring DR, Weichselbaum RR, Chen CT. Accurate 3D registration of CT, PET, and/or MR images of the brain. J Computer Assist Tomography. 1989;13(1):20–26. doi: 10.1097/00004728-198901000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Besl PJ, McKay ND. A Method for Registration of 3-D Shapes. IEEE Trans Pattern Analysis and Machine Intelligence. 1992;14(2):239–256. [Google Scholar]
- 8.Lazebnik RS, Lancaster TL, Breen MS, Lewin JS, Wilson DL. Volume registration using needle paths and point landmarks for evaluation of interventional MRI treatments. IEEE Trans Med Imaging. 2003;22(5):653–660. doi: 10.1109/TMI.2003.812246. [DOI] [PubMed] [Google Scholar]
- 9.Breen MS, Lancaster TL, Lazebnik RS, Nour SG, Lewin JS, Wilson DL. Three-dimensional method for comparing in vivo interventional MR images of thermally ablated tissue with tissue response. J Mag Res Imag. 2003;18:90–102. doi: 10.1002/jmri.10324. [DOI] [PubMed] [Google Scholar]
- 10.Wilson D, Breen MS, Lazebnik RS, Nour S, Lewin JS. Radiofrequency thermal ablation: 3D MR-histology correlation for localization of cell death in MR lesion images. Proc. Internat Symp Biomed Imaging; Arlington, VA: IEEE Press. 2004. [Google Scholar]
- 11.Zarow C, Kim TS, Singh M, Chui HC. A standardized method for brain-cutting suitable for both stereology and MRI-brain coregistration. J Neuorsci Methods. 2004;139:209–215. doi: 10.1016/j.jneumeth.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Meyer CR, Boes JL, Kim B, Bland PH, Zasadny KR, Kison PV, Koral K, Frey KA, Wahl RL. Demonstration of accuracy and clinical versatility of mutual information for automatic multimodality image fusion using affine and thin-plate spline warped geometric deformations. Med Image Analysis. 1997;1(3):195–206. doi: 10.1016/s1361-8415(97)85010-4. [DOI] [PubMed] [Google Scholar]