Fig. 1.
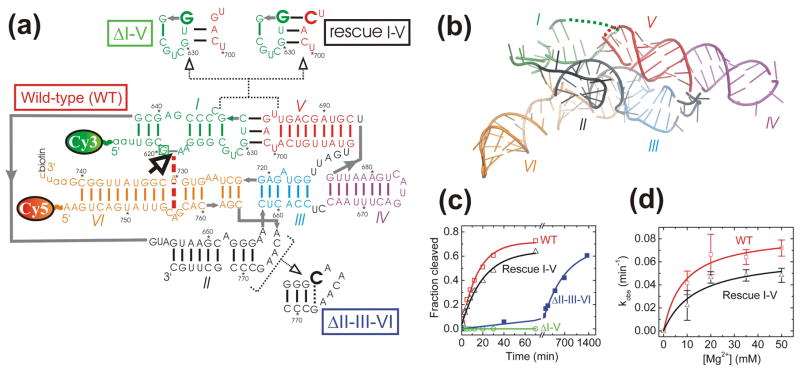
Structure and function of the well-studied G11 VS ribozyme that preserves the native VS RNA sequence. (a) Secondary structure depiction of the four VS ribozyme variants studied here, as based on two similar low-resolution structural models.18,20 Helices are color-coded, the bold open arrow next to the boxed G620 denotes the cleavage site, the red dashed line indicates a key interaction in the active site, gray lines denote direct backbone connectivities, and the Cy3, Cy5 and biotin labeling sites are indicated. Mutations in the ribozyme variants are shown next to heir names. All single molecule FRET experiments employed a 2′-O-methyl modification of G620 to render the substrate non-cleavable. (b) Tertiary structure model of the VS ribozyme (Zamel and Collins, unpublished data) using the same helix color code as in panel a. Some strand connectivities are incomplete in the model; the ones in the I-V kissing loop are indicated by dashed lines. (c) Cleavage time courses (data points) for each of the four ribozyme variants studied here, with single-exponential fits (lines) yielding the observed rate constants reported in the text. (d) Magnesium dependence of the observed cleavage rate constants of WT and Rescue I-V.