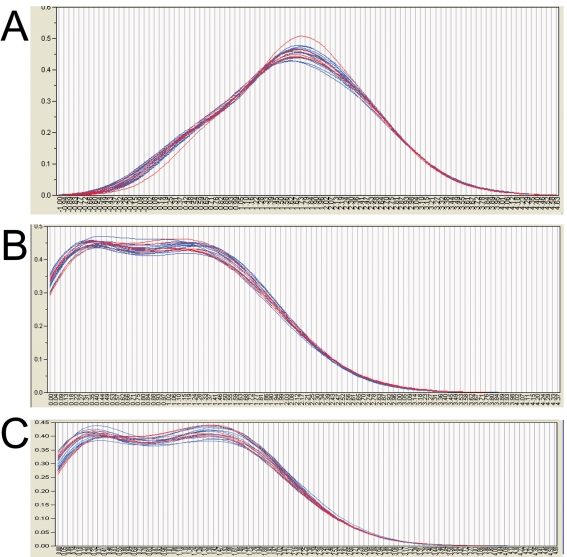
Figure 4. Overlayed kernel density estimates of Gene Expression by Array Hybridization and Sequence Read Frequencies.
14 SCZ samples are indicated by blue lines, and 6 control samples by red lines. The X-axis shows log transformed gene expression values while the Y-axis shows kernel densities. Panel A: Log10 transformed Affymetrix array hybridization signals. Panel B: Log10 transformed genome-aligned read frequencies. Panel C: Log10 transformed transcript-aligned read frequencies. Log10 transformed array hybridization values less than 1.69 (equivalent to a calibrated hybridization signal of 50) are considered noise. Without log transformation, samples showed greater variability in kernel densities and sequence read frequencies showed a near exponential decay (Fig. 1C, data not shown).