Abstract
Purpose
There are few genetic studies and clinical descriptions of Asian patients with X-linked ocular albinism (OA1). In the present study, the mutation analysis of G protein-coupled receptor 143 gene (GPR143) and clinical characteristics were assessed in Chinese patients with OA1.
Methods
Six families with OA1 were recruited from our pediatric and genetic eye clinic. Genomic DNA was prepared from venous leukocytes. The coding regions of GPR143 were amplified by polymerase chain reaction, and subsequently analyzed by direct sequencing. The variations detected were further evaluated in available family members as well as controls.
Results
Mutations in GPR143 were identified in each of the six families: c.849delT (p.Val284SerfsX15); c.238_240delCTC (p.Leu80del); c.658+1G>A, c.353G>A (p.Gly118Glu); g.1103_7266del6164 (p.Gly84AlafsX65), which resulted in a deletion of exons 2 and 3; and g.25985_26546del562 (p.Gly296ValfsX26), which resulted in a deletion of exon 8. Of these six, c.353G>A is a known mutation, while the other five are novel. All affected patients had nystagmus, poor visual acuity, and foveal hypoplasia. However, hypopigmentation of the iris and fundus was very mild in these patients.
Conclusions
Five novel mutations and one known mutation were identified in six Chinese families with OA1. These results expand the mutation spectrum of GPR143, and demonstrate the clinical characteristics of OA1 among the Chinese.
Introduction
X-linked ocular albinism (OA1; OMIM 300500), also called ocular albinism type 1, is the most common form of ocular albinism. Most affected white males exhibit nystagmus, poor visual acuity, iris translucency, foveal hypoplasia, an albinotic fundus, and normally pigmented skin and hair [1-4]. Other findings may include photophobia, strabismus, and misrouting of the optic tracts, resulting in loss of stereoscopic vision [5-7]. In some OA1 patients of black people, O'Donnell found hypopigmentation of the iris and retina may be subtle, such that these patients present a nonalbinotic fundus and little iris translucency [8]. However, the characteristics of OA1 have not been well defined in Asians. The misdiagnosis of OA1 as congenital nystagmus is not uncommon, and is usually due to lack of clinical experience or lack of phenotypic characterization for certain ethnic groups [9-14]. Such misdiagnoses might have gone undetected before the OA1 gene was identified.
OA1 is caused by mutations in the G protein-coupled receptor 143 (GPR143) gene (OMIM 300500), originally also known as the OA1 gene, located at Xp22.32 [15-17]. Various types of mutations in GPR143 have been identified in Caucasians living in the Netherlands, the United Kingdom, the United States, Germany, Canada, Belgium, France, Italy, and South Africa (albinism). Interestingly, genetic studies of Chinese patients with OA1 are rare.
In this study, sequence analysis of GPR143 resulted in the identification of mutations in each family, including five novel and one known mutations. Furthermore, we describe the clinical characteristics of OA1 patients and female carriers from six Chinese families.
Methods
Patients
There are 15 patients and 7 carriers participated in this study (Figure 1, Table 1), further more, 100 normal male controls mainly from the South of China also take part in, all these people are collected by our Pediatric and Genetic Clinic of Zhongshan Ophthalmic Center. Informed consent conforming to the tenets of the Declaration of Helsinki was obtained from each participant before the study. Medical and ophthalmic histories were obtained. Ophthalmological examination including visual acuity assessment , slit lamp biomicroscopic and ophthalmoscopic observation was performed by Drs. Guo and Zhang. Anterior and posterior segments of the eyes were documented by slit lamp and fundus photography. Flash visual evoked potential (VEP) was performed in one 4-year-old patient (proband in family 3). Genomic DNA was prepared from leukocytes of peripheral venous blood using the phenol-chloroform extracted method.
Figure 1.
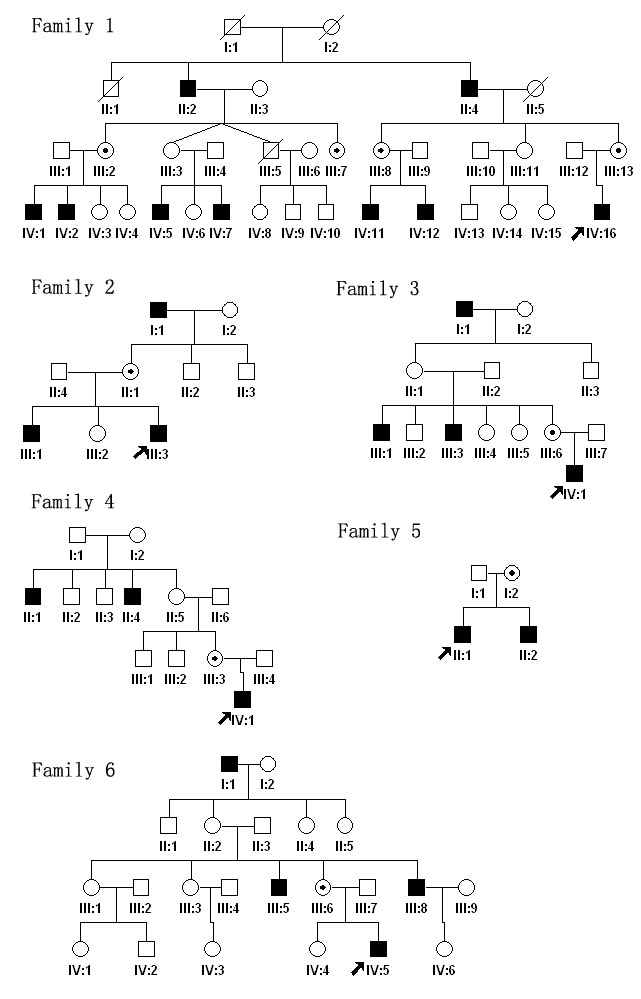
Pedigrees of the six families are shown. Black filled symbols indicate patients affected with OA1 in each family. Dot-marked symbols represent carriers. The proband is marked by arrow in each family.
Table 1. Summary of clinical findings in affected males and carriers.
| Family | ID#patients | Gender | Age (yrs) | First symptom | VA (corrected) | Iris hypopigmentation | Fundus a,b,c | Nys |
|---|---|---|---|---|---|---|---|---|
| 1 |
IV16 |
M |
8 |
nystagmus |
0.2/0.2 |
normal |
no,no, yes |
yes |
| |
IV5 |
M |
27 |
nystagmus |
0.25/0.3 |
mild |
no,no,yes |
yes |
| |
IV7 |
M |
23 |
nystagmus |
0.3/0.2 |
mild |
no,no,yes |
yes |
| |
IV2 |
M |
24 |
nystagmus |
0.2/0.25 |
normal |
no,no,yes |
yes |
| |
II4 |
M |
64 |
nystagmus |
0.4/0.2 |
normal |
no,yes,yes |
yes |
| |
II2 |
M |
77 |
nystagmus |
0.12/0.06 |
normal |
no,yes,yes |
yes |
| |
IV11 |
M |
11 |
nystagmus |
0.3/0.3 |
normal |
no,no,yes |
yes |
| |
IV12 |
M |
9 |
nystagmus |
0.2/0.12 |
normal |
no,no,yes |
yes |
| 2 |
III3 |
M |
4 |
poor VA |
0.1/0.1 |
mild |
ND |
yes |
| |
III1 |
M |
6 |
poor VA |
0.2/0.2 |
obvious |
no,no,yes |
yes |
| 3 |
IV1 |
M |
4 |
poor VA |
ND |
mild |
no,no,yes |
yes |
| 4 |
IV1 |
M |
7 |
nystagmus |
0.2/0.2 |
mild |
no,yes,yes |
yes |
| 5 |
II1 |
M |
12 |
nystagmus |
0.2/0.2 |
normal |
no,no,yes |
yes |
| |
II2 |
M |
7 |
nystagmus |
0.2/0.3 |
normal |
no,no,yes |
yes |
| 6 |
IV5 |
M |
4 |
nystagmus |
ND |
mild |
no,yes,yes |
yes |
| Carriers | ||||||||
| 1 |
III13 |
F |
38 |
NA |
0.8/1.0 |
normal |
no,no,no |
NA |
| |
III7 |
F |
41 |
NA |
0.8/1.0 |
normal |
no,no,no |
NA |
| |
III8 |
F |
36 |
NA |
1.0/1.0 |
normal |
no,no,no |
NA |
| |
III2 |
F |
50 |
NA |
1.0/1.0 |
normal |
no,no,no |
NA |
| 3 |
III6 |
F |
31 |
NA |
1.2/1.2 |
mild |
no,yes,no |
NA |
| 4 |
III3 |
F |
33 |
NA |
1.0/1.0 |
mild |
no,yes,no |
NA |
| 6 | III6 | F | 31 | NA | 1.0/1.0 | normal | no,yes,no | NA |
Clinical characteristics including gender, age, first symptom, visual acuity, iris, and fundus hypopigmentation form 15 OA1 patients and 7 carriers were conclude in this table. Abbreviations: albinotic fundus (a); hypopigmentation (b); foveal hypoplasia (c); not determined (ND); not applicable (NA), visual acuity(VA); nystagmus (Nys).
Mutation analysis
Eleven pairs of primers (Table 2) were used to amplify the nine coding exons and the adjacent intronic sequences of GPR143 (NCBI human genome build 36.2, GeneID 4935, NC_000023.9 for gDNA, NM_000273 for mRNA, NP_000264 for protein). Sequence analysis was performed as previously described [18]. The numbering system of the nucleotides for 404 amino acids, but not 424, was used for mutation description as previously described [19] (see in “In silico analysis” part).
Table 2. Primers for amplifying and sequencing GPR143 genomic segments.
| Exon | Forward primer (5′- 3′) | Reverse primer (5′- 3′) | Product size (bp) | Annealing temperature(°C) |
|---|---|---|---|---|
| 1A |
CTCCTCCGCCCGCCCAAGCATCAC |
CCCAGGCAGCCGAGAAGGTC |
464 |
68 |
| 1B |
CCGCGCCTAGGGACCTTCTGCT |
AACCCGCGGGCCTCTCGTCCTCAC |
399 |
70 |
| 2 |
CTTTCTTCCTTTTCCCTCCTTGTC |
GTTTGCTGCTGCTGCGATTTG |
360 |
58 |
| 3 |
CACGTGCGGCTTCCTGAC |
TTGGCCTCTTATAAAAATGA |
385 |
56 |
| 4 |
GGGCTTTCCTCTGTGTACATTTTC |
CCCTGAGACAACGGCCTAACC |
334 |
60 |
| 5 |
GCATTTCCCTTTTTGTTCTCATCC |
AGGCCTGCACATTTTCATTTATTG |
406 |
58 |
| 6 |
TTGCTTCCTGCCCCTCTGG |
ACTTGCTCCCCTGTCCTCTGT |
400 |
60 |
| 7 |
TGCACCTGGCCCTCTTAGTTTC |
TCAGGAGGCCAAGACAGAGGAT |
441 |
60 |
| 8A |
AAACCAACCCACCAACCAGTCAAC |
GCATGCTCAGGGCTTCGTCA |
395 |
60 |
| 8B |
CCAGCCCAGGGATTTCTCTT |
ACCCCGCCATGCACAGGAC |
329 |
60 |
| 9 |
AGCTGATGACAAACCTGCTAG |
CCCTTTCTCCTATCCTAAAG |
330 |
58 |
| FAM5 |
CCTAGGGACCGGATGGGACAAC |
CTCGCCAACAGTTACACAGCTC |
487 |
60 |
| FAM6 |
GTCCCCTTTCTTCCCTTCTCTACT |
GTGGTGTTTGTCTTTGGTTTTGTG |
297 |
60 |
| SSCP-1 |
AGCCACGCAGCTCGTGCTGAGCTTCCAGCC |
CCCAGGCGCTGATCAGATTCCAACCCGCGG |
250 |
70 |
| SSCP-2 |
CATCCTCTTATCTTGACTTCC |
CCCAGAGAGCTTCCCTAGT |
224 |
58 |
| SSCP-3 | CATGTTCTCTTTACCTGCTGC | AGTGAGACCTTGTCTCTGAAG | 227 | 56 |
Listed are primer sequences, sizes of PCR products, and annealing temperature used for the amplification. Primers 1-9 were used to amplify and sequence the GPR143 genimic segments. Primers FAM5 and FAM6 were used to detect the deletion boundaries in families 5 and 6, respectively. Primers SSCP-1 to 3 were used to amplify exons 1, 5, and 7 for heterduplex-SSCP analysis.
Any variations detected, except for two large intragenic deletions, were further evaluated in available family members as well as in 100 Chinese normal male controls using heteroduplex-single strand conformation polymorphism (SSCP) analysis, as previously described [20]. Three additional pairs of primers (Table 2, available on request) [21] were synthesized for heteroduplex-SSCP analysis of exons 1, 5, and 7.
In family 5, no PCR products were produced for the DNA fragments encompassing exons 2 and 3 of GPR143, suggesting a deletion in this region. To define the boundaries of the deletion, we designed a series of primer pairs based on sequences between exons 1 and 4 to amplify junctional fragments harboring the deletion. The deletion boundary was finally identified using the primer pairs shown in Table 2 (FAM5). In family 6, no PCR product was detected for exon 8. We used the same technique to find the deletion boundary with the primer pairs FAM6 shown in Table 2.
In silico analysis
Direct sequencing of the PCR products was performed with an ABI BigDye Terminator Cycle Sequencing Kit v3.1 (ABI Applied Biosystems, Foster City, CA) using an ABI3100 Genetic Analyzer. Sequencing results from patients as well as GPR143 consensus sequences from the NCBI human genome database (NM_000273) were imported into the SeqManII program of the Lasergene package (DNAStar Inc., Madison, WI) to identify variations. Each mutation was confirmed by bidirectional sequencing. Mutation descriptions followed the nomenclature recommended by the Human Genomic Variation Society (HGVS). To evaluate the splice site mutation, we used the Automated Splice Site Analysis (ASSA) server [22] to analyze the possible effects of this kind of mutation.
Results
Clinical phenotype
The clinical characteristics of OA1 in Chinese individuals are described as in Table 1 and Figure 2 and Figure 3. Table 1 shows the gender, age and clinical characteristics of the 15 patients and 7 carriers from the six families. Figure 2 and Figure 3 show some of the patients and carriers′ iris and fundus image respectively.
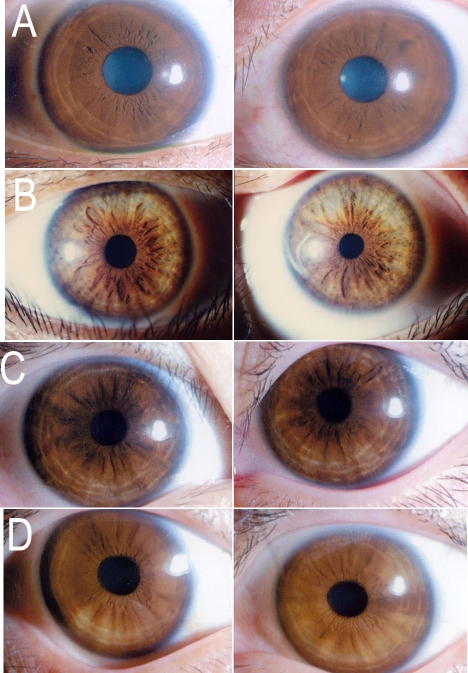
Figure 2.
Photographs of irises from normal control, patients, and carriers. Both of eyes′ irises are presented. A is a normal person whose irises exhibited normal pigmentation. B shows the irises of proband (III:3) in family 2. He is the only one patient who had obvious iris hypopigmentation. Obvious hypopigmentation is observed in both of eyes in picture B. C is the proband (IV:1) of family 4, showing slightly hypopigmentation in peripheral of the both eyes. D is carrier (III:3) in family 4. Her iris hypopigmentation is also very mild in both of eyes.
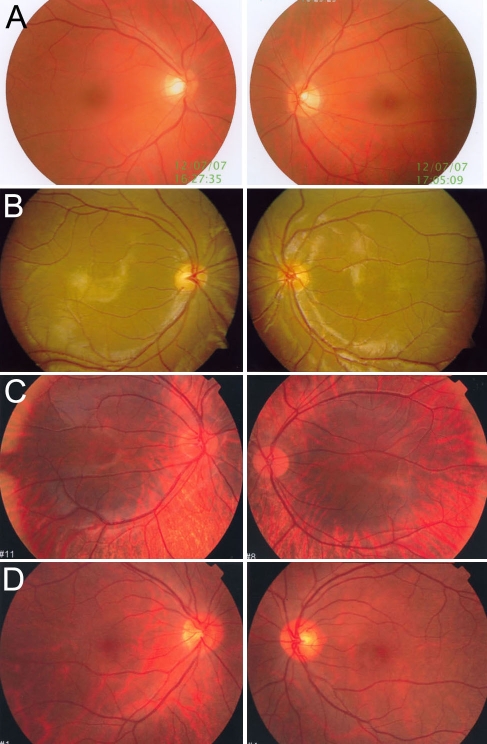
Figure 3.
Photographs of fundi from normal control, patients, and carriers. Both of eyes′ fundi are presented. A is the fundi from a normal control. B shows the fundi of proband (IV:16) in family 1. There is normal pigmentation and severe foveal hypoplasia in both of eyes in picture B. C presents the fundi of proband IV:1 from family 4. There was hypopigmentation in the posterior of the fundus and severe foveal hypoplasia in both of eyes in picture C. D shows the fundi from carrier III:3 in family 4. There was pigmentary mosaicism in the retinal pigment epithelium, and the fovea was normal in both of eyes.
Reduced visual acuity and nystagmus
All OA1 patients who came to our ophthalmic center had nystagmus and poor visual acuity as a first symptom. The nystagmus was present since birth, and corrected visual acuity (VA) was between 0.1–0.3. No VA data were obtained for families 3 and 6 because the patients were not old enough (Table 1) to cooperate with the examination. Flash VEP was performed for the family 3 proband, and no abnormalities were observed. The carriers did not show any symptoms, and their VA was normal (Table 1). All participants studied had normal skin and hair.
Iris hypopigmentation
This study examined 15 Chinese OA1 patients. Compared with normal individuals (Figure 2A), only one patient had obvious iris hypopigmentation (Figure 2B). Six patients exhibited mild peripheral iris hypopigmentation, as shown in Figure 2C, and eight patients had no abnormalities. Of the OA1 carriers, two exhibited mild peripheral iris hypopigmentation that could easily have been overlooked (Figure 2D), while the other carriers exhibited iris pigmentation that was similar to that in normal individuals.
Hypopigmentation in the fundus and foveal hypoplasia
This study examined the fundi of 13 patients'. Compared with normal individuals (Figure 3A), eight patients had normal pigmentation Figure 3B, three had mild hypopigmentation, and only two exhibited an albinotic fundus (Figure 3C). However, all of the patients had severe foveal hypoplasia (Table 1). In the proband of family 1 (Figure 3B), there was no typical albinotic fundus, and the fundus pigmentation had no abnormalities compared with normal individuals (Figure 3A). The only sign of the disease we observed was foveal hypoplasia. In the proband of family 4 (Figure 3C), hypopigmentation was apparent in the posterior of the fundus, and foveal hypoplasia was evident. Of the OA1 carriers, some had pigmentary mosaicism in the retinal pigment epithelium—for example, carrier III:3 in family 4 (Figure 3D)—while others had no hypopigmentation (Table 1).
Mutation analysis
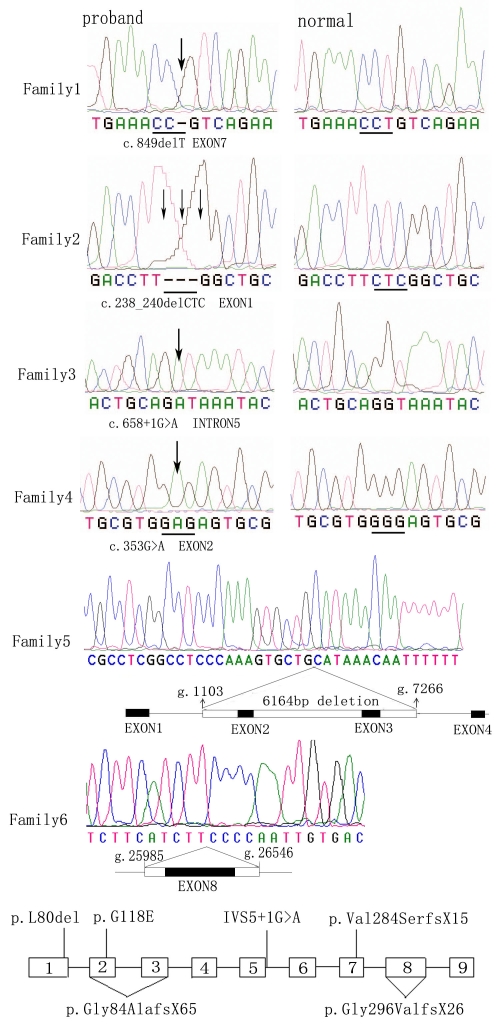
Sequence analysis of GPR143 detected six mutations among the six families (Table 3, Figure 1, and Figure, 4). Each of the six families studied had one of the following mutations: c.849delT (p.Val284SerfsX15); c.238_240delCTC (p.Leu80del); c.658+1G>A; c.353G>A (p.Gly118Glu); g.1103_7266del6164 (p.Gly84AlafsX65), which resulted in a deletion of exons 2 and 3; and g.25985_26546del562 (p.Gly296ValfsX26), which resulted in a deletion of exon 8. Of these six, c.353G>A is a known mutation, and the other five are novel. These mutations were present in affected hemizygous males and obligate heterozygous female carriers in the families, but were not detected in unaffected males and 100 controls by heteroduplex-SSCP analysis. The locations of the six mutations in GPR143 are shown in Figure 4.
Table 3. GPR143 mutations identified in six Chinese families with OA1.
| Patients | Nucleotide change | Exon/intron | Effect |
|---|---|---|---|
| Family 1 |
c.849delT |
e7 |
p.Val284SerfsX15 |
| Family 2 |
c.238_240delCTC |
e1 |
p.Leu80del |
| Family 3 |
c.658+1G>A |
i5 |
Loss of splicing acceptor |
| Family 4* |
c.353G>A |
e2 |
p.Gly118Glu |
| Family 5 |
g.1103_7266del6164 |
Loss of exons 2 and 3 |
p.Gly84AlafsX65 |
| Family 6 | g.25985_26546del562 | Loss of exons 8 | p.Gly296ValfsX26 |
Five novel and one known mutations were identified in six Chinese families. The mutation was named according to the nomenclature recommended by Human Genomic Variation Society (HGVS). In the third column, i5 stands for intron 5. The asterisk indicates that the mutation has been previously reported.
Figure 4.
Sequencing analysis of six families with OA1. The mutant GPR143 sequence (left) and corresponding normal sequence (right) are shown for families 1–4. The exact mutation is labeled under each sequence according to the nomenclature recommended by Human Genomic Variation Society (HGVS). Each mutation is marked with an arrow. In family 5, direct sequencing image was obtained by direct sequencing. The long band in Family 5 including exons 2 and 3, represents the large intragenic deletion region, which encompasses about 6,164 bp from g.1103 to g.7266 of GPR143. In family 6, direct sequencing image was obtained by direct sequencing, and the lower long band represents the deletion from g.25985 to g.26546 encompassing exon 8. The location of the six mutations within GPR143 is exhibited at the bottom of the Figure 4. Exons are shown as open boxes numbered 1–9, and thin lines represent introns. The locations of the six mutations found in this study are indicated beside the exons.
The c.849delT mutation (Figure 4, Table 3) is thought to induce a frameshift, creating a termination at codon 892. The resulting gene product lacks about a fourth of its amino acids, including the last transmembrane domain of the C-terminus. The c.238_240delCTC mutation resulted in a leucine deletion in the 80th amino acid position, and the c.353G>A mutation resulted in a replacement of glutamic acid with glycine in the 118th amino acid positions. The splice site mutation, c.658+1G>A, was expected to result in a loss of the corresponding splice sites (Figure 4, Table 3). The ASSA server [22] result predicted that the c.658+1G>A mutation would significantly change the strength of the splicing acceptor (from 9.2 bits to –3.6 bits) and would therefore abolish the natural splice site, corresponding to an approximately 594.6 fold decrease in the predicted affinity for this site. Two large intragenic deletions, g.1103_7266del6164 and g.25985_26546del562, resulted in the loss of an entire exon and produced what we believe to be a nonfunctional protein.
Discussion
In our study, we found five novel mutations and one known mutation in GPR143 among six unrelated Chinese families with OA1. All of the patients exhibited congenital nystagmus, poor visual acuity, mild hypopigmentation of the iris and fundus, and severe foveal hypoplasia.
Previous studies indicate that there is a higher detection of mutations in GPR143 in OA1 patients. Schiaffino et al. [2] screened the entire GPR143 coding sequence and detected mutations in one-third of patients. Schnur et al. [3] reported a higher frequency (90%) of mutations in their OA1 patient group. Here, our result indicate that Chinese OA1 patients also have a high frequency of mutations in GPR143 and further confirm that GPR143 is the major locus for OA1. Based on the high detection of mutations in GPR143, molecular genetic testing and prenatal diagnosis should be made available to Chinese OA1 patients.
Of the six mutations in our OA1 patients, there were four deletion mutations, one missense mutation, and one splice site mutation. These mutations mainly cluster within exons 1, 2, 3, and 7 of GPR143 (Figure 4), and the locations of the mutations are similar to previous studies. Large intragenic deletion is a common mutation in GPR143. Bassi et al. [23] found a diverse prevalence of large deletions between European (<10%) and North American (>50%) patients with OA1. Interestingly we also found two large intragenic deletions in our OA1 patients. These results may indicate that large intragenic deletions are also common in Chinese OA1 patients. The c.353G>A (p.Gly118Glu) mutation, resulting in the replacement of glutamic acid with glycine at the 118th amino acid position, has been identified in Caucasian populations [3] and patients from the Netherlands [23]. And this is the third time to report it, the mutation appeared in different ethnic groups indicate that c.353G>A (p.Gly118Glu) may be a hot spot mutation. Our result also shows that Chinese OA1 patients may have a similar spectrum of mutation in GPR143 compared with the White group. This mutation may therefore be a hot spot mutation in different ethnic groups. The findings from our study show that Chinese OA1 patients may have a similar spectrum of mutation in GPR143 compared with the Caucasian group.
The OA1 patients in our study exhibited congenital nystagmus and poor visual acuity with mild hypopigmentation of the iris and fundus. Liu et al. [12] reported a large X-linked recessive inheritance in a Chinese family with nystagmus as a prominent and consistent manifestation of the phenotype. None of the patients in the family had the complete classical phenotype of ocular albinism, and patients were initially misdiagnosed with congenital nystagmus. The disease gene was mapped to a region approximately 10.6 Mb in size, flanked by DXS996 and DXS7593 on Xp22 with a significant peak multipoint LOD score. Analysis of 21 candidate genes in the region revealed a novel p.S89F mutation in the second transmembrane domain of GPR143.
In our OA1 patients, iris hypopigmentation ranged from mild to normal, with the exception of one patient. It was difficult to distinguish any differences compared to normal individuals. Our OA1 patients showed little change in clinical characteristics of the fundus, exhibiting only mild hypopigmentation and sometimes even normal pigmentation. Severe foveal hypoplasia was the prominent clinical feature observed in Chinese OA1 patients, which may be a useful criterion for distinguishing OA1 from nystagmus of unknown cause. As hypopigmentation of the iris and fundus is not typical among the Chinese, OA1 could easily be misdiagnosed as another disease due to the presence of nystagmus and severely reduced visual acuity. Although foveal hypoplasia may be a key indicator of OA1, unawareness of the clinical characteristics of OA1 and lack of experience might explain why OA1 is rarely reported in the Chinese population. Here, we describe the characteristic signs of OA1 in Chinese patients, potentially facilitating the diagnosis of OA1 in this population. For patients presenting with nystagmus and reduced visual acuity, careful evaluation of iris color, fundus color, and foveal development is critical. Furthermore, molecular genetic testing of GPR143 might be useful for establishing a diagnosis of OA1.
In summary, this report identified five novel mutations and one known mutation in GPR143 of Chinese OA1 patients. Moreover, we examined the clinical features of Chinese OA1 patients and found that foveal hypoplasia may be a key indicator of OA1 in patients with nystagmus of unknown cause. Identification of mutations in GPR143 and more information regarding clinical manifestations should facilitate early diagnosis, appropriate early therapy, and genetic counseling for this disease.
ACKNOWLEDGMENTS
We thank the patients and family members for their participation. This study was supported in part by the National 863 Plan of China (04AA104092; to X.G.), the National Natural Science Foundation of China (30572006; to Q.Z.), the National Science Fund for Distinguished Young Scholars of China (30725044; to Q.Z.), and the Guangdong Natural Science Foundation (04009335; to X.G.).
References
- 1.Creel D, O'Donnell FE, Jr, Witkop CJ., Jr Visual system anomalies in human ocular albinos. Science. 1978;201:931–3. doi: 10.1126/science.684419. [DOI] [PubMed] [Google Scholar]
- 2.Schiaffino MV, Bassi MT, Galli L, Renieri A, Bruttini M, De Nigris F, Bergen AA, Charles SJ, Yates JR, Meindl A. Analysis of the OA1 gene reveals mutations in only one-third of patients with X-linked ocular albinism. Hum Mol Genet. 1995;4:2319–25. doi: 10.1093/hmg/4.12.2319. [DOI] [PubMed] [Google Scholar]
- 3.Schnur RE, Gao M, Wick PA, Keller M, Benke PJ, Edwards MJ, Grix AW, Hockey A, Jung JH, Kidd KK, Kistenmacher M, Levin AV, Lewis RA, Musarella MA, Nowakowski RW, Orlow SJ, Pagon RS, Pillers DA, Punnett HH, Quinn GE, Tezcan K, Wagstaff J, Weleber RG. OA1 mutations and deletions in X-linked ocular albinism. Am J Hum Genet. 1998;62:800–9. doi: 10.1086/301776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen B, Orlow SJ. The ocular albinism type 1 gene product is an N-glycoprotein but glycosylation is not required for its subcellular distribution. Pigment Cell Res. 2001;14:485–90. doi: 10.1034/j.1600-0749.2001.140609.x. [DOI] [PubMed] [Google Scholar]
- 5.Walker BA, Martyn LJ, Coffman T. X-linked ocular albinism. Birth Defects Orig Artic Ser. 1971;7:200–2. [PubMed] [Google Scholar]
- 6.Valle O. Ocular albinism. Duodecim. 1982;98:950–7. [PubMed] [Google Scholar]
- 7.Salati R, Magni R, Musolino M, Nucci P, Polenghi F. Electronystagmographic investigation in X-linked ocular albinism. Ophthalmic Genet. 1997;18:209–15. doi: 10.3109/13816819709041436. [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell FE, Jr, Green WR, Fleischman JA, Hambrick GW. X-linked ocular albinism in Blacks. Ocular albinism cum pigmento. Arch Ophthalmol. 1978;96:1189–92. doi: 10.1001/archopht.1978.03910060023005. [DOI] [PubMed] [Google Scholar]
- 9.Preising M, Op de Laak JP, Lorenz B. Deletion in the OA1 gene in a family with congenital X linked nystagmus. Br J Ophthalmol. 2001;85:1098–103. doi: 10.1136/bjo.85.9.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faugere V, Tuffery-Giraud S, Hamel C, Claustres M. Identification of three novel OA1 gene mutations identified in three families misdiagnosed with congenital nystagmus and carrier status determination by real-time quantitative PCR assay. BMC Genet. 2003;4:1. doi: 10.1186/1471-2156-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sallmann GB, Bray PJ, Rogers S, Quince A, Cotton RG, Carden SM. Scanning the ocular albinism 1 (OA1) gene for polymorphisms in congenital nystagmus by DHPLC. Ophthalmic Genet. 2006;27:43–9. doi: 10.1080/13816810600677834. [DOI] [PubMed] [Google Scholar]
- 12.Liu JY, Ren X, Yang X, Guo T, Yao Q, Li L, Dai X, Zhang M, Wang L, Liu M, Wang QK. Identification of a novel GPR143 mutation in a large Chinese family with congenital nystagmus as the most prominent and consistent manifestation. J Hum Genet. 2007;52:565–70. doi: 10.1007/s10038-007-0152-3. [DOI] [PubMed] [Google Scholar]
- 13.Guo X, Li S, Jia X, Xiao X, Wang P, Zhang Q. Linkage analysis of two families with X-linked recessive congenital motor nystagmus. J Hum Genet. 2006;51:76–80. doi: 10.1007/s10038-005-0316-y. [DOI] [PubMed] [Google Scholar]
- 14.Lam BL, Fingert JH, Shutt BC, Singleton EM, Merin LM, Brown HH, Sheffield VC, Stone EM. Clinical and molecular characterization of a family affected with X-linked ocular albinism (OA1). Ophthalmic Genet. 1997;18:175–84. doi: 10.3109/13816819709041432. [DOI] [PubMed] [Google Scholar]
- 15.Schnur RE, Trask BJ, van den Engh G, Punnett HH, Kistenmacher M, Tomeo MA, Naids RE, Nussbaum RL. An Xp22 microdeletion associated with ocular albinism and ichthyosis: approximation of breakpoints and estimation of deletion size by using cloned DNA probes and flow cytometry. Am J Hum Genet. 1989;45:706–20. [PMC free article] [PubMed] [Google Scholar]
- 16.Schnur RE, Nussbaum RL, Anson-Cartwright L, McDowell C, Worton RG, Musarella MA. Linkage analysis in X-linked ocular albinism. Genomics. 1991;9:605–13. doi: 10.1016/0888-7543(91)90353-g. [DOI] [PubMed] [Google Scholar]
- 17.Winship IM, Babaya M, Ramesar RS. X-linked ocular albinism and sensorineural deafness: linkage to Xp22.3. Genomics. 1993;18:444–5. doi: 10.1006/geno.1993.1495. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q, Li S, Xiao X, Jia X, Guo X. The 208delG mutation in FSCN2 does not associate with retinal degeneration in Chinese individuals. Invest Ophthalmol Vis Sci. 2007;48:530–3. doi: 10.1167/iovs.06-0669. [DOI] [PubMed] [Google Scholar]
- 19.Huang L, Sadler L, O'Riordan MA, Robin NH. Delay in diagnosis of Williams syndrome. Clin Pediatr (Phila) 2002;41:257–61. doi: 10.1177/000992280204100410. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Minoda K, Zeng R, Wu Z, Xiao X, Li S, Zhang F. Exon-by-exon screening for RB germline mutations using Heteroduplex-SSCP analysis. Yan Ke Xue Bao. 1997;13:5–11. [PubMed] [Google Scholar]
- 21.Mayeur H, Roche O, Vetu C, Jaliffa C, Marchant D, Dollfus H, Bonneau D, Munier FL, Schorderet DF, Levin AV, Heon E, Sutherland J, Lacombe D, Said E, Mezer E, Kaplan J, Dufier JL, Marsac C, Menasche M, Abitbol M. Eight previously unidentified mutations found in the OA1 ocular albinism gene. BMC Med Genet. 2006;7:41. doi: 10.1186/1471-2350-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P, Guo X, Jia X, Li S, Xiao X, Zhang Q. Novel mutations of the PAX6 gene identified in Chinese patients with aniridia. Mol Vis. 2006;12:644–8. [PubMed] [Google Scholar]
- 23.Bassi MT, Bergen AA, Bitoun P, Charles SJ, Clementi M, Gosselin R, Hurst J, Lewis RA, Lorenz B, Meitinger T, Messiaen L, Ramesar RS, Ballabio A, Schiaffino MV. Diverse prevalence of large deletions within the OA1 gene in ocular albinism type 1 patients from Europe and North America. Hum Genet. 2001;108:51–4. doi: 10.1007/s004390000440. [DOI] [PubMed] [Google Scholar]