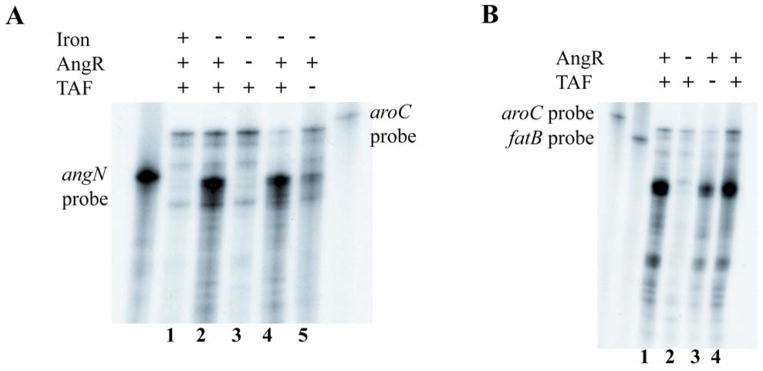
Fig. 6.
Effect of iron, AngR and TAF on transcription of angN and fatB. A. Ribonuclease protection assay using a probe specific for the angN gene and total RNA isolated from cultures grown under iron-rich and iron-limiting conditions. V. anguillarum wild type (lanes 1, 2 and 4); V. anguillarum AngR-deficient strain (lane 3); V. anguillarum TAF-deficient strain (lane 5). RNA obtained under: iron-rich conditions, lane 1 (2 μg/ml ferric ammonium citrate); iron-limiting conditions, lanes 2 and 4 (1.5 μM EDDA) and lanes 3 and 5 (0.5 μM EDDA). B. Ribonuclease protection assay with the same RNA used in panel A from the cultures grown under iron-limiting conditions using a fatB-specific riboprobe. V. anguillarum wild type (lanes 1 and 4); V. anguillarum AngR-deficient strain (lane 2); V. anguillarum TAF-deficient strain (lane 3). The aroC-specific riboprobe is included in both Ribonuclease protection assays to provide an internal control for the quality of the RNA and the amount of RNA loaded onto the gel, since the aroC gene is expressed independently of the iron concentration of the cell.