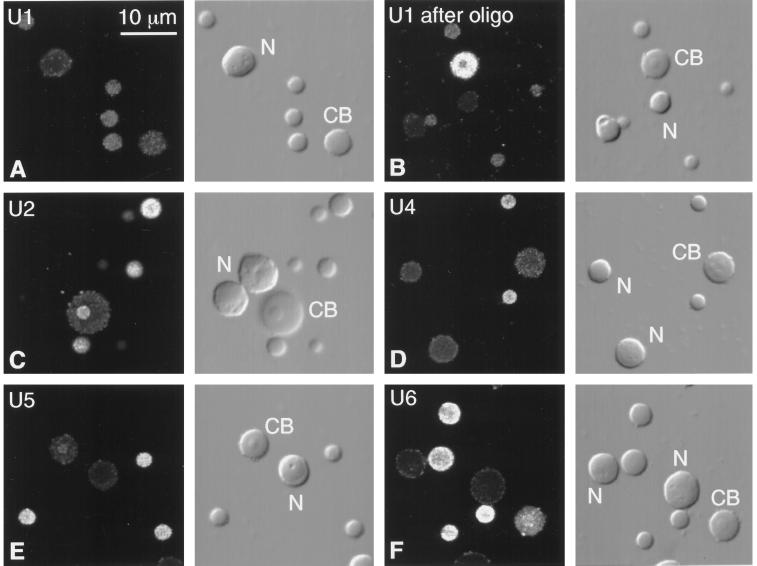
Figure 3.
Fluorescent in situ hybridization of splicing snRNAs (U1, U2, U4, U5, and U6) in Cajal bodies, B-snurposomes, and nucleoli from spread GV preparations of Xenopus. Each pair of panels shows the fluorescent image on the left and the corresponding DIC image on the right. CB, Cajal body; N, nucleolus. (A) U1 is weakly detectable in B-snurposomes, but the Cajal body and nucleoli are barely above the background level of unhybridized controls. (B) Truncated U1 accumulates in Cajal bodies of oocytes treated with a deoxyoligonucleotide that removes the cap and first 20 nucleotides from the 5′ end. B-snurposomes and nucleoli are similar to those in untreated oocytes. (C) U2 is strong in B-snurposomes and easily detectable in the matrix of the Cajal body. (D) U4 is strong in B-snurposomes, but the Cajal body and nucleoli are at background level. (E) U5 is strong in B-snurposomes and detectably above background in the matrix of the Cajal bodies. (F) U6 is strong in B-snurposomes and gives the strongest reaction of all the splicing snRNAs in the matrix of the Cajal bodies.