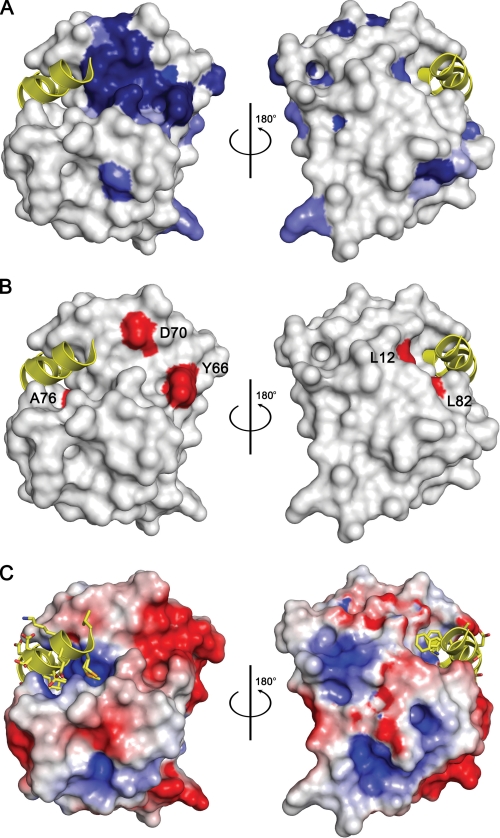
FIGURE 2.
Residue conservation and Phd-binding site of Doc. A, surface representation of DocH66Y. Residues that are conserved in Doc sequences are colored blue with the intensity of the color reflecting the degree of conservation: darkest blue represents full conservation in all sequences, and lightest blue represents 50% conservation (residue conservation calculated according to Livingstone and Barton (39) and based upon the sequence alignment shown in supplemental Fig. S1). Two views are shown 180° apart. The bound fragment of Phd is shown as a yellow helix ribbon. B, mapping of toxicity-eliminating mutations on the surface of Doc. Residues that lead to a non-toxic form of P1 Doc (19) are colored red and labeled. Asp-60 and Tyr-66 are part of the conserved sequence motif. Leu-12 and Leu-82 are within the Phd52–73Se-binding site. However, the corresponding mutations L12P and L82P are likely to disrupt helices α1 and α4, respectively, in agreement with the observation that both lead to a protein that loses its toxicity as well as its regulatory activity. The orientations are the same as in panel A. C, electrostatic surface potential mapped on the surface of Doc. Negatively charged regions are colored red, and positively charged regions are colored blue. The bound fragment of Phd is shown as a yellow helix ribbon. The orientations are the same as in panel A.