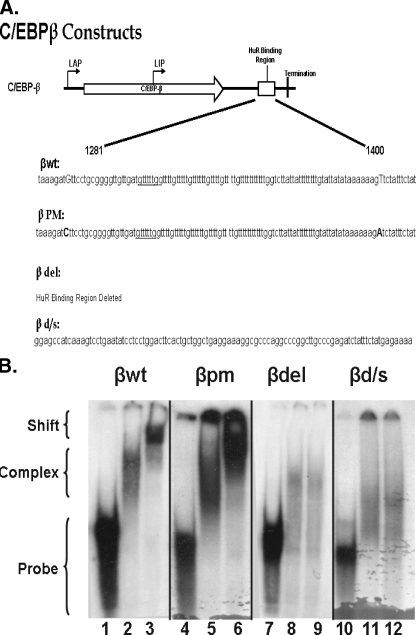
FIGURE 1.
RNA gel shift analysis of HuR binding to wild type and mutant C/EBPβ 3′-UTR AREs. A, a schematic of C/EBPβ mRNA indicating the approximate translation initiation sites for liver activating protein 1 and 2 (LAP) and liver inhibitory protein (LIP) forms of C/EBPβ, the HuR binding domain, and termination of translation is shown. The graphic below describes the sequence alterations in the mutants. In βwt, the uppercase bases indicate the sequence altered to form βpm, in which the uppercase bases indicate the mutations used to create BglII restriction sites in βwt. βdel is a religation of the construct after removal of the BglII fragment. βd/s consists of removal of the BglII fragment and insertion of a 101-base fragment that does not bind HuR. B, cytosolic extracts and a radiolabeled probe corresponding to the ARE (βwt and βpm), βd/s, or the region flanking (βdel) the C/EBPβ 3′-UTR were used to perform RNA gel shift and super shift analysis. Lanes 1, 4, 7, and 10, probe alone; lanes 2, 5, 8, and 11, probe plus 10 μg of adipocyte cytosolic extract; lanes 3, 6, 9, and 12, supershift of complex formed as in lanes 2, 5, 8, and 11 using the 3A2 anti-HuR monoclonal antibody. The gel shifts shown were performed at the same time, and separation was achieved on two separate gels, βwt andβdel on one andβpm andβd/s on the second. The arrangement was for logical presentation. These data are representative of two individual gel shifts with each probe as the alterations were made, using two individual preparations of cytosolic extracts.