Abstract
A triple-locus nucleotide sequence analysis based on toxin regulatory genes tcdC, tcdR and cdtR was initiated to assess the sequence variability of these genes among Clostridium difficile isolates and to study the genetic relatedness between isolates. A preliminary investigation of the variability of the tcdC gene was done with 57 clinical and veterinary isolates. Twenty-three isolates representing nine main clusters were selected for tcdC, tcdR, and cdtR analysis. The numbers of alleles found for tcdC, tcdR and cdtR were nine, six, and five, respectively. All strains possessed the cdtR gene except toxin A-negative toxin B-positive variants. All but one binary toxin CDT-positive isolate harbored a deletion (>1 bp) in the tcdC gene. The combined analyses of the three genes allowed us to distinguish five lineages correlated with the different types of deletion in tcdC, i.e., 18 bp (associated or not with a deletion at position 117), 36 bp, 39 bp, and 54 bp, and with the wild-type tcdC (no deletion). The tcdR and tcdC genes, though located within the same pathogenicity locus, were found to have evolved separately. Coevolution of the three genes was noted only with strains harboring a 39-bp or a 54-bp deletion in tcdC that formed two homogeneous, separate divergent clusters. Our study supported the existence of the known clones (PCR ribotype 027 isolates and toxin A-negative toxin B-positive C. difficile variants) and evidence for clonality of isolates with a 39-bp deletion (toxinotype V, PCR ribotype 078) that are frequently isolated worldwide from human infections and from food animals.
Clostridium difficile, a gram-positive spore-forming strictly anaerobic bacillus, is responsible for 15 to 25% of postantibiotic diarrhea and 95% of pseudomembranous colitis in humans. C. difficile is the main cause of infectious nosocomial diarrhea in adult patients and colitis during or following an antibiotic treatment (3, 4, 28). Since 2003, severe infections in hospitalized patients have increased in North America (United States and Canada and particularly the province of Quebec) (26, 27, 33, 34) and more recently in some European countries (England, Wales, Ireland, The Netherlands, Belgium, Luxembourg, and France) (18, 21, 22, 51). Most of these epidemics led to the isolation of quinolone-resistant strains sharing common characteristics (PCR ribotype 027, toxinotype III, and strains producing, besides the toxins TcdA and TcdB, the binary toxin CDT). C. difficile infection is also associated with enteric diseases in animals, including horses, dogs, and swine (5, 19, 37, 45). Its implication in food-borne diseases and as a zoonotic agent have been recently demonstrated (38, 43, 44).
The major virulence factors of C. difficile are the large clostridial toxins TcdA and TcdB, which are produced by all pathogenic strains isolated from patients suffering from postantibiotic diarrhea with C. difficile infection. TcdA and TcdB toxin-encoding genes together with tcdR (formerly tcdD [39]) and tcdC, which regulate toxin production positively and negatively, respectively, are located on the chromosome within a pathogenicity locus named PaLoc (31, 32). A third toxin, the binary toxin CDT, whose encoding genes cdtA and cdtB are located outside the PaLoc, is produced by some strains. A recently identified gene, cdtR, positively regulates the production of CDT (8). The cdt locus (CdtLoc) is the region containing cdtR, cdtA, and cdtB, and only those isolates that had the tcdA and tcdB genes carried cdtR (8). Ninety-six percent of cdtR-positive isolates also carry the cdtAB genes (the full-length CdtLoc encoding the functional binary toxin) or at the very least, fragments of these genes (a truncated CdtLoc) (8). The CdtLoc is widely disseminated; apart from toxin A-negative toxin B-positive (A− B+) isolates, which do not harbor CdtLoc, more than 90% of C. difficile isolates analyzed contain this region (8). CDT-producing strains are found mainly in outbreaks and represent less than 10% of isolates. In most cases, they are variant strains with changes in the PaLoc region (40). The role of CDT in the pathogenesis and the disease is not yet known but must be considered as an additional virulence factor. CDT-producing strains were most often isolated from severe cases of C. difficile colitis (1).
Recently, the discovery of various deletions in the tcdC gene gave rise to numerous hypotheses about the increased virulence of PCR ribotype 027 strains in humans (33, 47, 53). These strains harbored a 1-bp deletion at position 117 that was always associated with an 18-bp deletion (29). Some other tcdC genotypes with or without deletions have also been described (9, 29, 47).
The purposes of this study were, first, to study the variability of tcdC gene sequences from C. difficile isolates originating from various hosts (humans and animals) and harboring various types of deletion (as determined by gel electrophoresis) or no deletion (wild type [wt]) in tcdC, and second, to undertake a triple-locus sequence analysis based on the toxin regulatory genes tcdC, tcdR, and cdtR to assess the sequence variability of these genes and then to study the genetic relatedness of the isolates. We demonstrate that the tcdR, tcdC, and cdtR genes have evolved divergently among most strains studied, except in isolates harboring a 39-bp or a 54-bp deletion in tcdC.
MATERIALS AND METHODS
Bacterial isolates.
Fifty-three C. difficile isolates chosen according to their hosts and the estimated (by gel electrophoresis) tcdC deletion size have been studied (Table 1), including 25 isolates with a 36- or 39-bp deletion (7 from horses, 4 from piglets, and 14 from humans). The other human isolates consisted of six with a 54-bp deletion; nine belonging to the 027 clone; three with an 18-bp deletion, belonging to toxinotype III; and two A− B+ toxin variants (from France, toxinotype VIII). In addition, eight isolates without a deletion in tcdC (four from French human patients and four from German horses) were included. Among the French 027 isolates, the “historical” 027 C. difficile strain AIP 196.84 was included. This strain was isolated in 1984 by M. R. Popoff at the Institut Pasteur from a young patient with severe pseudomembranous colitis (36). The reference strain VPI 10463 and strain 630 were also included. Purity of strains was controlled by streaking on meat-yeast extract agar medium complemented with 5% egg yolk.
TABLE 1.
Origins, sequence types, binary toxin results, and correspondence of strains examined in this study
| Strain/sequencea | tcdC gene type by deletion (bp) or toxinb | tcdC allele (positions 1 to 600)c |
tcdC genotypes corresponding to the study byd:
|
ST no. (allelic profile for tcdC, tcdR, cdtR)e | Binary toxin CDT | Hostf | Country | |
|---|---|---|---|---|---|---|---|---|
| Spigaglia and Mastrantonio (46) | Curry et al. (9) | |||||||
| AIP 236.05* | 18 | C | ST6 (C, A, A) | + | Human | France | ||
| AIP 274.03* | 18 | C | ST6 (C, A, A) | + | Human | France | ||
| AIP 284.03* | 18 | C | ST6 (C, A, A) | + | Human | France | ||
| AIP 141.07* | 36 | J | sc-16 | ST3 (J, D, A) | + | Human | France | |
| AIP 278.03* | 36 | J | sc-16 | ST4 (J, D, B) | + | Human | France | |
| AIP 10229* | 39 | A | A type | A type | ST1 (A, C, E) | + | Horse | Germany |
| AIP 10234 | 39 | A | A type | A type | + | Horse | Germany | |
| AIP 10235 | 39 | A | A type | A type | + | Horse | Germany | |
| AIP 10236 | 39 | A | A type | A type | + | Horse | Germany | |
| AIP 10239 | 39 | A | A type | A type | + | Horse | Germany | |
| AIP 10250* | 39 | A | A type | A type | ST1 (A, C, E) | + | Horse | Switzerland |
| AIP 10251 | 39 | A | A type | A type | + | Horse | Switzerland | |
| AIP 115.04 | 39 | A | A type | A type | + | Pig | France | |
| AIP 119.04 | 39 | A | A type | A type | + | Pig | France | |
| AIP 142.05 | 39 | A | A type | A type | + | Human | France | |
| AIP 176.05 | 39 | A | A type | A type | + | Human | France | |
| AIP 180.04* | 39 | A | A type | A type | ST1 (A, C, E) | + | Pig | France |
| AIP 199.07 | 39 | A | A type | A type | + | Human | France | |
| AIP 23.07 | 39 | A | A type | A type | + | Human | France | |
| AIP 24.07* | 39 | A | A type | A type | ST1 (A, C, E) | + | Human | France |
| AIP 331.06 | 39 | A | A type | A type | + | Human | France | |
| AIP 334.06 | 39 | A | A type | A type | + | Human | France | |
| AIP 336.06 | 39 | A | A type | A type | + | Human | France | |
| AIP 406.06 | 39 | A | A type | A type | + | Pig | France | |
| AIP 410.06 | 39 | A | A type | A type | + | Human | France | |
| AIP 414.06 | 39 | A | A type | A type | + | Human | France | |
| AIP 49.03 | 39 | A | A type | A type | + | Human | France | |
| AIP 89.07 | 39 | A | A type | A type | + | Human | France | |
| AIP 10245* | 54 | B | ST2 (B, F, D) | + | Human | France | ||
| AIP 10246 | 54 | B | + | Human | France | |||
| AIP 10247 | 54 | B | + | Human | France | |||
| AIP 10248 | 54 | B | + | Human | France | |||
| AIP 162.05* | 54 | B | ST2 (B, F, D) | + | Human | France | ||
| AIP 329.06* | 54 | B | ST2 (B, F, D) | + | Human | France | ||
| AIP 10249* | Δ117 + 18-bp | I | sc-1 | ST5 (I, A, A) | + | Human | France | |
| AIP 16.07 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| AIP 196.84* | Δ117 + 18-bp | I | sc-1 | ST5 (I, A, A) | + | Human | France | |
| AIP 330.06 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| AIP 339.06 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| AIP 4.07 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| AIP 421.06 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| AIP 427.06 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| AIP 449.06 | Δ117 + 18-bp | I | sc-1 | + | Human | France | ||
| CM000287* | Δ117 + 18-bp | I | sc-1 | ST5 (I, A, A) | + | Human | Canada | |
| CM00041* | Δ117 + 18-bp | I | sc-1 | ST5 (I, A, A) | + | Human | Canada | |
| AIP 10228* | Wild type | H | sc-9 | ST10 (H, B, C) | − | Horse | Germany | |
| AIP 10230 | Wild type | H | sc-9 | − | Horse | Germany | ||
| AIP 10237 | Wild type | G | − | Horse | Germany | |||
| AIP 10241 | Wild type | G | − | Horse | Germany | |||
| AIP 11.07 | Wild type | G | − | Human | France | |||
| AIP 333.06* | Wild type | C | sc-13 | ST6 (C, A, A) | + | Human | France | |
| AIP 418.06* | Wild type | E | ST7 (E, B, C) | − | Human | France | ||
| AIP 81.07 | Wild type | F | − | Human | France | |||
| AM180355* | Wild type | H | sc-9 | ST9 (H, A, C) | − | Human | Switzerland | |
| VPI 10463* | Wild type | G | ST8 (G, A, C) | − | Human | |||
| AIP 145.05* | Wild type (A− B+) | D | sc-7 | † | − | Human | France | |
| AIP 192.05* | Wild type (A− B+) | D | sc-7 | † | − | Human | France | |
*, strains/sequences were tested for tcdC, tcdR, and cdtR.
The bp values 18, 36, 39, and 54 are sizes of deletions in the tcdC gene; Δ117 + 18-bp indicates a 1-bp deletion at position 117, associated with an 18-bp deletion in the tcdC gene; Wild type, a tcdC gene without deletion.
cdtR genes of toxin A-negative toxin B-positive C. difficile isolates could not be amplified. They were tcdC allele D and tcdR allele E.
tcdC gene types corresponding to those found by Spigaglia and Mastrantonio (46) and Curry et al. (9) of the 57 strains examined in this study.
†, positions relative to the ATG start codon of the tcdC sequence from strain VPI 10463.
All human strains were obtained from diseased people.
Sequencing of toxin regulatory genes tcdC, tcdR and cdtR.
Bacteria were cultivated in Trypticase yeast extract glucose (TGY) broth in an anaerobic atmosphere for 18 h. For DNA extraction, two protocols were used: (i) purification of nucleic acids with InstaGene matrix (Bio-Rad) according to the manufacturer's instructions; and (ii) FTA cards (Whatman). Briefly, bacteria were cultured in TGY broth in an anaerobic atmosphere for 18 h. Sixty-five microliters was applied to the card and left to dry. Discs were punched out of the sample area, placed in a tube, and washed three times with 200 μl of FTA purification reagent (Whatman). Washed discs were allowed to dry completely. A disc was added directly into the PCR mixture.
PCRs were performed in a final volume of 60 μl containing 0.5 μM concentrations of forward and reverse primers, 200 μM concentrations of each deoxynucleoside triphosphate, and 3 U of Taq DNA polymerase (Invitrogen) in a 1× amplification buffer containing 1.5 mM MgCl2.
Unless otherwise stated, the DNA template for all PCRs was a disc prepared with FTA. The entire tcdC gene was amplified by PCR using primers C1 and C2 (47). The cycling conditions were as follow: denaturation at 94°C for 45 s; annealing at 50°C for 45 s; extension at 72°C for 45 s (40 cycles); and a final extension at 72°C for 10 min. A 473-bp fragment of the tcdR gene was amplified by PCR by using primers PAL11 and Pal12 and conditions described by Spigaglia and Mastrantonio (46). For this PCR, 5 μl of DNA prepared with Instagene matrix was added to 55 μl of mixture. The cdtR gene was amplified by PCR using primers P1605 forward (5′-AGCATAAATATACCTTAATTCTAACTATC-3′) and P1510 reverse (5′-TCTTGAGACATCTCCTTTTTCT-3′). The cycling conditions were as follows: denaturation at 94°C for 45 s; annealing at 50°C for 45 s; extension at 72°C for 75 s (35 cycles); and a final extension at 72°C for 10 min. PCR products were purified and sequenced (both strands) with forward and reverse primers by MWG Biotech, Roissy CDG (France). Sizes of the analyzed internal fragments were 600 bp for tcdC (positions 1 to 600, relative to the ATG start codon), 385 bp for tcdR (positions 92 to 476 of the coding sequence), and 720 bp for cdtR (positions 1 to 720, relative to the GTG start codon).
Internal fragments of the cdtA and cdtB genes coding for the two peptidic chains of the binary toxin CDT were amplified using the cdtA and cdtB primers as described by Stubbs et al. (50). After a denaturation step at 94°C for 5 min, reactions were subjected to 30 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min.
Data analyses.
Genome sequences available (whole sequence or shotguns) were also included: strain 630 with a wt tcdC gene (whole sequence, GenBank accession no. AM180355) and two Canadian 027 isolates (strains QCD-32g58 and QCD-66c26) (shotguns, GenBank accession no. CM000287 and CM00041, respectively). The tcdC, tcdR, and cdTR gene sequences identified from strain VPI 10463 served as templates for retrieving the tcdC, tcdR and cdtR sequences from genomic or shotgun sequences.
Sequences were aligned by using a BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenetic analysis was conducted using Molecular Evolutionary Genetics Analysis (MEGA) software (version 3.1) (http://www.megasoftware.net) (23). The phylogenetic inference was based on the neighbor-joining distance method (41). Gene trees were constructed with the neighbor-joining method, using the Kimura two-parameter model (20) and bootstrapping algorithms contained in MEGA software (23). Each gene sequence that differed by one or more nucleotides was considered to be a different allele, and an arbitrary letter designation was assigned. Each unique allelic pattern over all three loci examined was identified as a sequence type (ST). Allelic profiles and sequence data were imported into the START software (version 2) (17) (http://www.mlst.net) to determine the mean G+C%. The average frequencies of synonymous substitutions per potential synonymous site (dS) and nonsynonymous substitutions per potential nonsynonymous site (dN) were also calculated with the START software to test the degree of selection on a locus. A 1,705-bp (for 21 strains) nucleotide composite sequence (derived for three concatenated gene fragments) was also aligned with MEGA version 3.1 software (http://www.megasoftware.net). Repeats within genes were searched with Tandem Repeats Finder software at http://tandem.bu.edu/trf/trf.html (6).
Cytotoxicity assay.
The presence of large clostridial toxins (mainly cytotoxin TcdB) was assessed by a cytotoxicity assay (35). Cells were cultivated in Dulbecco's modified Eagle medium supplemented with 5% fetal bovine serum. Vero (African green monkey kidney) cells were transferred into the wells of a 96-well Falcon tissue culture plate (Becton Dickinson Labware, Oxnard, CA) and grown for 24 h to form monolayers. Supernatants from 48-h cultures (optical density at 600 nm [OD600] ± standard deviation, 1.7 ± 0.2) in TYH broth (without glucose) inoculated with 100 μl of an overnight culture (OD600, 1.6 ± 0.2), incubated in an anaerobic atmosphere at 37°C, were used. Serial twofold dilutions of supernatants in the cell culture medium (100 μl, final volume) were added to the monolayers. The cells were monitored for 24 h and 48 h after incubation for morphological alteration. The cytotoxicity titer corresponds to the reciprocal of the greater dilution giving morphological alterations in 50% of the cells.
CDTa assay.
CDTa was assayed by in vitro ADP-ribosylation in 50 mM triethanolamine buffer (pH 7.5) containing 5 mM MgCl2, 10 mM dithiothreitol, 10 mM thymidine, 5 × 105 cpm of [32P]NAD (specific activity, 3,000 Ci mmol−1; PerkinElmer Life Sciences). Supernatants from 48-h cultures (OD600, 1.6 ± 0.2) in TGYH broth (with glucose) inoculated with 100 μl of an overnight culture (OD600, 1.7 ± 0.2), incubated in an anaerobic atmosphere at 37°C, were used. For in vitro ADP-ribosylation, 5 μl of C. difficile culture supernatant or a threefold serial dilution in Tris 50 mM (pH 7.5), 1 mg/ml bovine serum albumin was added to 5 μl of ADP-ribosylation buffer containing 2 μg of nonmuscular actin (Cytoskeleton). The reaction mixture was incubated for 1 h at 37°C, stopped by the addition of 7 μl of the sample buffer, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The gel was dried and processed on a phosphorimager (Typhon; Amersham Biosciences) for quantification of the ADP-ribosylated actin band.
Nucleotide sequence accession numbers.
Nucleotide sequences of the internal fragment genes analyzed in this work were deposited in the GenBank database under accession numbers EU271773 to EU271778 and EU271785 (tcdR); EU271779 to EU271783 (cdtR); and EU271784 (the tcdC gene sequence for strain AIP 162.05 with a 54-bp deletion in tcdC).
RESULTS
Allelic variation in the three toxin regulatory genes. (i) tcdC.
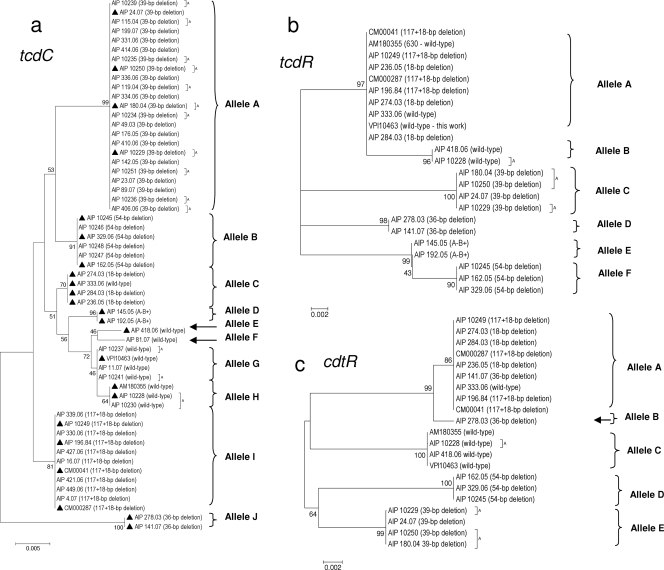
When all tcdC nucleotide sequences (600 nucleotides) were aligned, using BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), in comparison with the tcdC sequence from strain VPI 10463 (GenBank accession no. X92982), some recurrent discrepancies appeared all along the aligned sequences. Therefore, we decided to sequence the tcdC gene from strain VPI 10463, maintained in our culture collection (equivalent to CIP 109240 in the Collection of Bacteria of the Institut Pasteur). The tcdC sequence of this strain differed from the GenBank X92982 sequence by the insertion of three nucleotides and the replacement of two nucleotides within positions 442 to 454. These changes were fully in agreement with those found previously by Spigaglia et al. (47) and Curry et al. (9). Therefore, the updated sequence of our strain VPI 10463 will be used as the reference for tcdC. Among the 57 tcdC sequences studied, 43 (75.4%) presented deletions in tcdC. Four deletion sizes were encountered, 18 bp (at positions 330 to 347) alone or associated with a 1-bp deletion at position 117 (herein called Δ117 + 18-bp), 36 bp (at positions 301 to 336), 39 bp (at positions 341 to 379), and finally 54 bp (at positions 313 to 366). Ten alleles were identified (A to J, showing 1 to 20 nucleotide differences) (Fig. 1a and 3). Sequences with the same deletion (Δ117 + 18-bp, 36 bp, 39 bp, and 54 bp) clustered together (100% identity) and corresponded to alleles I, J, A, and B, respectively. Sequences with an 18-bp deletion clustered together with a strain without a deletion in tcdC (strain AIP 333.06; 100% identity, allele C). tcdC sequences without deletions (herein called wt tcdC) clustered in four close clusters (alleles E to H, showing 1 to 5 nucleotide differences). tcdC sequences from A− B+ toxin variants clustered together (100% identity, allele D).
FIG. 1.
Dendrograms showing genetic relationships of the C. difficile isolates based on allele sequences of individual toxin regulatory loci tcdC (a), tcdR (b), and cdtR (c). Fifty-seven isolates are shown for tcdC. From these 57 isolates, 23 isolates were selected (▴) for tcdR and cdtR analysis. Twenty-one isolates are shown for the cdtR locus (lack of amplification for the two A− B+ isolates). Dendrograms were reconstructed from the nucleotide sequence of each gene by using the neighbor-joining method. The genetic distances were computed by using the Kimura two-parameter model. The scale bar indicates the genetic distance. The number shown next to each node indicates the percent bootstrap value of 1,000 replicates. A, animal isolate.
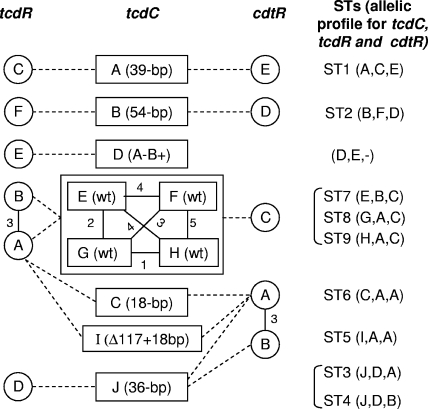
FIG. 3.
Diagram showing associations between tcdC, tcdR, and cdtR alleles among strains studied. The tcdC alleles are boxes A to J (midcolumn of the diagram) with the type of deletion in the tcdC gene (or the toxin type for the A− B+ toxin variants) noted in parentheses. Four close tcdC alleles, E, F, G and H, grouped in a box correspond to a wt tcdC gene. Open circles are tcdR alleles (A to F, left column). Gray circles are cdtR alleles (A to E, right column). Circles or boxes linked by a solid line represent close alleles; numbers beside the solid line represent nucleotide differences between alleles. Dashed lines represent relationships between the tcdC, tcdR and cdtR alleles. Corresponding STs are noted at the right of the diagram. tcdR and cdtR sequences were not determined for the unique tcdC F allele strain (AIP 81.07).
Most sequences with deletions of >1-bp (occurring in the region 301 to 376) had an upstream mutation or a 1-bp deletion that could lead to shorter predicted protein sequences. The deletion at position 117 (found alone or associated with an 18-bp deletion) creates a shift in the reading frame that introduces a TAG stop codon at position 196 that could lead to a predicted truncated protein (65 residues) (9). tcdC sequences harboring a 36-bp deletion had a mutation, G191A, that creates a TGA stop codon at position 190 and could lead to a predicted truncated amino acid sequence (63 residues). tcdC sequences harboring a 39-bp or a 54-bp deletion had a mutation, C184T, that creates a TAA stop codon at position 184 and could lead to a predicted truncated amino acid sequence (61 residues). Numerous tcdC genotypes have been described. Spigaglia and Mastrantonio (47) described 3 tcdC types (A, B, and C) (47), and Curry et al. (9) described 15 additional genotypes. Correspondence between the tcdC alleles defined in this study and the previously described tcdC genotypes is given Table 1. Allele A in this study corresponded to the tcdC type A described by Spigaglia and Mastrantonio (47), whereas alleles B, C, E, and F did not correspond to any described genotype.
It is noteworthy that in our study, all sequences harboring a deletion in tcdC were from CDT-producing isolates (as evidenced by amplification of internal fragments of cdtA and cdtB genes). An isolate with a wt tcdC gene also possessed CDT genes (strain AIP 333.06).
Clustering of predicted amino acid sequences from nucleotide sequences did not extensively modify the classification. All clusters remained unchanged, except for alleles C, E, G, and H (which contained the most nondeleted sequences) which clustered in a single group (data not shown). These results on tcdC sequences showed that for most strains (a few exceptions exist), a given combination of mutations in the sequence is strictly associated with a type of deletion (36 bp, 39 bp, and 54 bp).
(ii) tcdR.
To study the variability of the tcdR and cdtR genes, 23 isolates or sequences representative of the main clusters obtained from the tcdC gene analysis were selected (Fig. 1b). Fragments (385-bp sequences from positions 92 to 476 relative to the ATG start codon) were extracted from amplified tcdR sequences and then aligned and clustered. Six clusters corresponding to six tcdR alleles (A to F, with 2 to 14 nucleotide differences) were obtained (Fig. 1b and 3). Within each cluster, sequences had 100% identity. Clustering of predicted amino acid sequences from tcdR nucleotide sequences did not at all modify the classification (data not shown).
(iii) cdtR.
All but two cdtR sequences from the 23 selected isolates or sequences were amplified by PCR (the cdtR genes from the two A− B+ toxin variants could not be amplified). Fragments (720-bp sequences from positions 1 to 720 relative to the GTG start codon) were extracted, aligned, and clustered. Five clusters (alleles A to E, with 3 to 22 nucleotide differences) were obtained.
It is important to note that all cdtR allele E sequences, all from isolates with a 39-bp deletion in tcdC, had a G322T mutation that introduced a TAA stop codon at position 322 in place of a “GAA” codon. The resultant predicted protein could be truncated: 107 amino acids in place of 249 amino acids for an uninterrupted sequence. The exact role of this putative truncated CdtR regulator protein in the in vitro production of binary toxin CDT should be assessed. Clustering of predicted amino acid sequences from the cdtR nucleotide sequences did not at all modify the classification. All clusters remained unchanged (data not shown).
Genetic variation.
The internal gene fragments analyzed ranged from 385 bp (tcdR) to 720 bp (cdtR) (Table 2). The average G+C% was estimated to range from 20.3% for tcdR to 29.4% for tcdC. For comparison, the average G+C content for the whole genome of strain 630 is 29.1% (42). The average number of alleles per locus for the three genes was 7.0 (range, 5 to 10). The number of variable sites in a given locus ranged from 23 (tcdR) to 36 (cdtR). With respect to the very different lengths of the tcdR, tcdC and cdtR genes, the percentages of variable sites were somewhat close (5, 6, and 5%, respectively). The range of nucleotide differences between alleles for the three genes studied is given in Table 2. In tcdC, there was a continuum of values from 1 to 20 nucleotide differences, but it can be noted that allele J (36-bp deletion in tcdC) was divergent from other alleles with nucleotide differences ranging from 13 to 20 (data not shown). In tcdR, alleles E and F differed by only two nucleotides and alleles A and B by three nucleotides; other interallele nucleotide differences ranged from 7 to 14 nucleotides. In cdtR, alleles A and B differed by only one nucleotide; other alleles had 15 to 22 nucleotide differences. The average dN/dS values are shown in Table 2. All values were <1.0, indicating that most of the sequence variability identified is selectively neutral.
TABLE 2.
Genetic variations in the three sequenced loci
| Gene | No. of sequences | Fragment size (bp) | No. of alleles | No. of polymorphic sites (%) | No. of synonymous base substitutionsa | Range of nucleotide differences between alleles (% range) | Mean % of G+C content | dN/dSb |
|---|---|---|---|---|---|---|---|---|
| tcdC | 57 | 600 | 10 | 30 (5.0) | 10 | 1-20 (0.2-3.0) | 29.4 | 0.1875 |
| tcdR | 23 | 385 | 6 | 23 (6.0) | 4 | 2-14 (0.5-3.6) | 20.3 | 0.1697 |
| cdtR | 21 | 720 | 5 | 36 (5.0) | 17 | 3-22 (0.4-3.0) | 22.9 | 0.2709 |
Synonymous base substitutions are nucleotide changes which did not result in amino acid changes.
dN/dS, average frequencies of synonymous substitutions per potential synonymous site (dS) over nonsynonymous substitutions per potential nonsynonymous site (dN) were calculated with START software to test the degree of selection on a locus.
Triple-locus sequence typing—association between the tcdC, tcdR and cdtR alleles.
Multilocus sequence typing (MLST) is based on allelic differences in the nucleotide sequences of housekeeping or virulence-associated genes among bacterial strains (30). MLST distinguishes strains based on the observed variations in the nucleotide sequences of several loci rather than the degree of sequence variation in any single gene or locus. From allelic profiles of the three loci, 10 STs among the 21 isolates were identified (Fig. 2 and 3). Of these, six (60%) were identified once (ST3, ST4, ST7 to ST10). Each couple of STs, ST3 and ST4, ST5 and ST6, and ST8 and ST9, as well as ST9 and ST10 differed by a variation at a single locus. The four ST1 isolates were from animals (two horses and one piglet) and from a human and had a 39-bp deletion in tcdC. The four ST5 isolates were PCR ribotype 027 isolates (including three recent epidemic isolates from Quebec, Canada, and France and the French historic strain AIP 196.84). ST2 was represented by four recent human isolates from different hospitals and locations in France and had a 54-bp deletion in tcdC. All but one ST6 isolate had an 18-bp deletion. The remaining ST6 isolate (strain AIP 333.06) had a tcdC sequence similar to the other three isolates but had no deletion. The A− B+ toxin variants that could not be included in the composite sequence analysis and the triple-locus sequence analysis (no amplification of cdtR) were tcdC allele D and tcdR allele E, both alleles not encountered in other strains of the study.
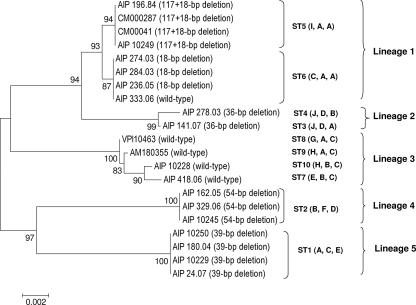
FIG. 2.
Dendrogram showing genetic relationships of 21 C. difficile isolates based on the composite sequence (1,705 bp) of three toxin regulatory genes, tcdC, tcdR, and cdtR. Two A− B+ toxin isolates were not included since their cdtR fragments could not be amplified. The dendrogram was obtained by the neighbor-joining method with the Kimura two-parameter model. Bootstrap values were calculated from 1,000 resamplings of the original data. ST, sequence type number. Alleles for tcdC, tcdR, and cdtR are shown in parentheses. A, animal isolate.
tcdC analysis showed that sequences with a given deletion clustered together and had 100% identity within a cluster. Sequences with a wt tcdC (no deletion) clustered into four close clusters (one to five nucleotide differences). When we examined tcdR analysis results with respect to the type of deletion in tcdC, we observed that isolates with tcdC deletion types of 36 bp, 39 bp, and 54 bp clustered separately (tcdC alleles D, C, and F, respectively). The two A− B+ toxin variants clustered together (allele E). Finally, a branch encompassed the rest of the isolates with two separate clusters: the largest (allele A) intermixed 10 sequences with different tcdC types (wt tcdC, with an 18-bp deletion and a Δ117 + 18-bp deletion), and the smallest cluster (allele B) contained two isolates with a wt tcdC gene.
For cdtR, the largest cluster (allele A) gathered sequences with different tcdC types: Δ117 + 18bp deletion type, 18-bp deletion alone, an isolate with a 36-bp deletion, and an isolate with a wt tcdC gene. Close to allele A, allele B (three nucleotide differences) was represented by a single isolate (AIP 278.03) with a 36-bp deletion. Alleles C, D, and E included sequences with wt tcdC and with a 54-bp deletion and a 39-bp deletion in tcdC, respectively. Some alleles (tcdC alleles A and B, tcdR alleles C and F, and cdtR alleles E and D) were found exclusively in the following combinations of alleles (i.e., sequence type or allelic profile for tcdC, tcdR, and cdtR): ST1 (alleles A, C, and E) and ST2 (alleles B, F, and D). These two STs were composed of strains harboring 39-bp and 54-bp deletions in tcdC, respectively. In these two groups of strains, the three toxin regulator genes could have coevolved. In contrast, tcdR allele A was found in strains with different tcdC alleles (wt tcdC, 18-bp deletion alone or associated with a 1-bp deletion at position 117, 36-bp deletions in tcdC). This fact was quite surprising, since the tcdC and tcdR genes are located within the same PaLoc. For cdtR, the situation is rather different, since all isolates with a wt tcdC gene (except AIP 333.06) clustered together (cdtR allele C) and separately from all isolates harboring an 18-bp deletion (associated or not with a 1-bp deletion at position 117; allele A) or a 36-bp deletion (allele B, with 20 nucleotides difference between alleles A and C). This clearly demonstrated that the evolutions of the three toxin regulatory genes were different, according to the genotype of tcdC.
Composite sequence analysis.
To determine the overall divergence of the sequenced fragments of the three loci studied, these sequences were spliced together to obtain a concatenated composite sequence for each isolate (1,705 bp). Due to the absence of the cdtR amplification product, the two A− B+ toxin variants were not included. Thus, 21 composite nucleotide sequences were aligned with MEGA software. For calculating the percentage of identity or divergence, in-frame insertions and deletions were not taken into account. The identity between the 21 composite sequences was found to be between 97.3 and 100%. An unrooted phylogenetic tree was constructed using the neighbor-joining approach (Fig. 2). The resultant clustering showed five lineages among six clusters. Lineage 1 was formed by two subclusters in which all but one of the isolates had an 18-bp deletion in tcdC associated or not with a 1-bp deletion at position 117. The remaining isolate (AIP 333.06) had a wt tcdC gene but was binary toxin CDT positive, like the other strains of the group. All these sequences differed by no more than three nucleotides. Lineage 3 contained four isolates with the wt tcdC gene. Lineages 2, 4, and 5 contained isolates with deletions of 36 bp, 54 bp, and 39 bp in tcdC, respectively. Overall, the clustering generated from composite sequence analysis was congruent with associations of alleles for the three genes studied.
Tandem repeat analysis.
Repeated sequences that code for eight-amino-acid repeats were previously identified in the tcdC gene (14, 47). It is well known that tandem repeats (TRs) may be responsible for recA-independent intragenic homologous recombination that leads to deletion or duplication of repeats by slipped-strand mispairing (7, 52). We examined the different alleles of the three regulatory genes for TRs by using Tandem Repeats Finder software. In tcdC, all TRs were located within positions 278 to 388, a region where all known deletions have been identified (positions 301 to 379). There were four TRs in wt tcdC alleles (E, F, G, and H), three TRs in the A− B+ toxin variant (allele D), two TRs in alleles with an 18-bp deletion (alleles C and I), one TR in the allele with a 36-bp deletion (allele J), and no TRs at all in alleles with the largest deletions, i.e., alleles A (39-bp deletion) and B (54-bp deletion). In wt tcdC alleles, the four TRs had period sizes of 18, 18 (two different TRs of 18-bp found), 27, and 36 bp; copy numbers ranging from 2.9 to 4.2; and percent matches between copies ranging from 62 to 82%. TRs were rather rare in tcdR (allele D, one TR, period size of 69 bp) and cdtR (allele D, one TR of period size 57 bp; and allele E, two TRs of period sizes 33 bp and 57 bp).
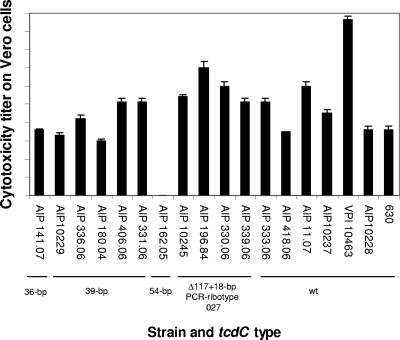
Toxin assays. (i) Large clostridial toxins.
Production of large clostridial toxins (mainly TcdB) was assessed with 18 strains in a cytotoxicity assay of Vero cell monolayers by testing supernatants of 48-h cultures done in a medium without glucose (TYH broth). Results are shown in Fig. 4. All but one strain tested produced large clostridial toxins, as evidenced by cytotoxicity on Vero cells. The negative strain AIP 162.05 possessed the tcdA and tcdB genes as detected by PCR. After a 48-h incubation, titers ranged from 1,000 (strain AIP 180.04) to 9,548,000 (reference strain VPI 10463 with a wt tcdC gene). PCR ribotype 027 isolates (including the historic strain AIP 196.84) had cytotoxicity titers ranging from 128,000 to 2,048,000.
FIG. 4.
Cytotoxicity of 48-h C. difficile cultures on monolayer Vero cells. Isolates of various tcdC types: wt (no deletion), 18-bp deletion associated with a 1-bp deletion at position 117 (characteristic of the PCR ribotype 027 clone), 36-bp, 39-bp, and 54-bp deletions were tested by a cytotoxicity assay. The high-level-toxin-producing reference strain VPI 10463 (wt tcdC gene) was included. The cytotoxicity titer corresponds to the greater dilution giving a morphological alteration in 50% of the cells. Values are averages of duplicate cultures, and bars indicate standard errors.
(ii) Binary toxin CDT.
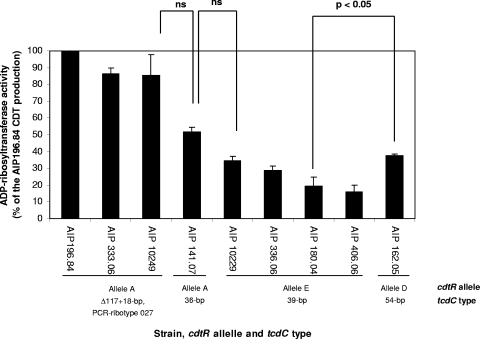
Production of binary toxin CDT was assessed by an ADP-ribosyltransferase assay of nine strains harboring the cdtA and cdtB genes, as evidenced by PCR (Fig. 5). All strains tested demonstrated an ADP-ribosyltransferase activity. When ADP-ribosyltransferase activity was normalized against serial dilutions of a CdtA recombinant protein, a quantitative ratio of 1:5 was found between lower and higher CDT producers (not shown). Among the four cdtR allele A strains tested, the reference strain AIP 196.84 (PCR ribotype 027) produced the largest amount of CDT, confirming previous results (35, 36). Then, ADP-ribosyltransferase activity results were expressed as percentages of the activity of strain AIP 196.84. Both of the 027 strains AIP 10249 and AIP 333.06 had about 80% activity. It is noteworthy that the fourth cdtR allele A strain studied (AIP 141.07, a non-027 strain) had a lower level of activity (50%, nonsignificant) than 027 isolates (Fig. 5). The remaining strains (cdtR alleles D and E) had between 15 and 33% activities. cdtR allele E strains, for which a stop codon was found in the cdtR sequence, had lower activities that might or might not be significantly different from strains without a stop codon in cdtR (alleles A and D) (Fig. 5).
FIG. 5.
ADP-ribosyltransferase activities of 48-h C. difficile cultures. CDTa was assayed in isolates harboring various tcdC types. The quantification of the ADP-ribosylated band on a gel corresponded to the ADP-ribosyltransferase activity. ADP-ribosyltransferase activity results were expressed on the graph as percentages of the activity of the high-level-CDT-producing strain AIP 196.84 (100% activity). Values are averages of duplicate cultures, and bars indicate standard errors. Statistical analyses of logarithmic ADP-ribosyltransferase activity values were calculated using analysis of variance and Bonferroni post hoc compensation for multiple comparisons. P values for comparison of activities of strains AIP 10249 to AIP 141.07, AIP 141.07 to AIP 10229, and AIP 180.04 to AIP 162.05 are indicated. ns, nonsignificant.
DISCUSSION
The aim of this study was to study the variability of the tcdC gene in C. difficile isolates from various hosts and then to develop a triple-locus sequence analysis based on three toxin regulatory genes, tcdC, tcdR, and cdtR, to assess the relatedness of the isolates.
The tcdC sequences accumulated deletions of various sizes (18 bp, 36 bp, 39 bp, and 54 bp), whereas the tcdR and cdtR genes were never found deleted (in 23 and 21 strains studied, respectively). TRs are known to be responsible for recA-independent homologous recombination within genes, leading to deletion or duplication of repeats mainly by slipped-strand mispairing (7, 52). Within the tcdC alleles (except alleles A [39-bp deletion in tcdC] and B [54-bp deletion in tcdC] for which no TR was detected), all TRs were located within positions 277 to 388 of the gene, a region where all deletions are already known to have occurred. Among the sizes of the different TRs identified in tcdC (18 bases, 27 bases, and 36 bases; copy numbers ranging from 2.1 to 5.2), all but one size (27-base TRs) corresponded to frequently observed deletion sizes. The wt tcdC alleles (E to H) and A− B+ toxin variants (allele D) accumulate TRs (three to four copies). The frequency of homologous recombination within tcdC is probably low and needs to be evaluated, but some examples that could support this hypothesis may be noted. A few sequences in the literature and in this work shared the same profile of mutations in tcdC but differed only by the presence of a deletion. Curry et al. (9) described type sc-8, whose sequence is identical to the sc-1 type encountered in all PCR ribotype 027 isolates, except for a G92A mutation and an 18-bp deletion. It is not excluded that types sc-8 and sc-1 were derived after a recombination event. This hypothesis could not be ruled out, since both types were isolated in the same hospital. The type B tcdC sequence described by Spigaglia and Mastrantonio (47) and the tcdC sequence from VPI 10463 had 100% identity, except for an 18-bp deletion in the former. In our study, strain AIP 333.06, which had no deletion in tcdC, belonged to sequence type 6, a sequence type found exclusively in all three strains with an 18-bp deletion (Fig. 2).
The evidence for various deletions in the tcdC gene raises numerous hypotheses about the increased production of toxins TcdA and TcdB by PCR ribotype 027 strains in humans (33, 47, 53). Matamouros et al. (32) have shown that the hypertoxigenicity phenotype of epidemic 027 strains might not be due to the 18-bp in-frame deletion in tcdC and concluded that the 1-bp deletion at position 117 leading to a stop codon is certainly at least in part responsible for this phenotype. Our cytotoxin experiments confirmed that strain VPI 10463 with a wt tcdC gene produced the highest level of toxins. Among the four 027 isolates tested, the historic strain AIP 196.84 produced the highest level of toxins (titer, 1:9,548,000). The production of cytotoxins was rather variable among 027 isolates and was not significantly different from most non-27 isolates with a deleted or a wt tcdC gene, except for the high-level-producing strain VPI 10463 (Fig. 4). These observations indicate that other factors may affect the level of production of large clostridial toxins such as, for example, the nutritional sensor CodY (12). cdtR, a positive regulator of binary toxin production, was recently identified (8). Among nine strains tested for binary toxin production, the 027 isolates (cdtR allele A) had the higher ADP-ribosyltransferase activity. A non-027 cdtR allele A strain (AIP 141.07) had a lower activity (nonsignificant difference). The truncated positive regulator CdtR discovered in allele E isolates did not seem to dramatically alter the production of binary toxin, since their ADP-ribosyltransferase activities were close to those found in other cdtR alleles without a stop codon (alleles A and D) (Fig. 5). Correlation between the CDT-positive regulator cdtR allele and level of CDT production should be confirmed with a greater number of strains. The exact roles of the different putative truncations in toxin regulatory proteins TcdC and CdtR on the production of large clostridial toxins (TcdA and TcdB) and binary toxin CDT, respectively, should also be determined to assess whether these changes participate to increase the virulence of strains.
All strains except the A− B+ toxin variants possessed a cdtR gene, even if they contained truncated cdtA and cdtB genes (49). All but one CDT-positive isolate harbored a deletion in the tcdC gene in our study. cdtR analysis showed that all CDT-negative strains contained a wt tcdC gene and clustered separately from isolates with an 18-bp or an 36-bp deletion type in tcdC (except for strain AIP 333.06). Finally, isolates with 39-bp and 54-bp deletions in tcdC were clearly separated from other strains by tcdC, tcdR, and cdtR clustering (Fig. 3).
The triple-locus sequence analysis, as well as the composite sequence analysis, showed that deletion in the tcdC gene may be used as a mark of clonality, since isolates with a given deletion in the tcdC (18 bp, associated or not with a 1-bp deletion at position 117, 39 bp, and 54 bp) gene were grouped together in the same cluster and in the same sequence type. The two strains with a 36-bp deletion in tcdC had identical tcdC and tcdR alleles but differed only by a nucleotide in their cdtR genes. In contrast, strains with a wt tcdC gene were found to be more heterogeneous, since they clustered in four groups and four STs (one to five nucleotide differences).
In our study, three groups of strains had specific alleles in tcdC, tcdR, and cdtR that were not encountered elsewhere. These three genes might have coevolved. The first group is constituted by A− B+ Clostridium difficile toxin variants (tcdC allele D and tcdR allele E; no amplification of cdtR; as described above). tcdC allele D and tcdR allele E were not identified in other strains of the study. The A− B+ toxin variant strains belong to toxinotype VIII and serogroup F (10, 11, 16) and PCR ribotype 017. These A− B+ toxin variants did not have intact or truncated CDT loci and did not have the cdtR gene, the entire cdt locus being replaced by a conserved 68-bp sequence (8). The second group was represented by isolates harboring a 39-bp deletion in tcdC (ST1, binary toxin CDT positive, tcdC allele A). In the preliminary study, tcdC allele A (a 39-bp deletion type, from 11 animal isolates and 12 human isolates) has 100% identity with the tcdC type A described by Spigaglia and Mastrantonio (47). They were all toxinotype V, with similar PCR ribotype 078 profiles. Our results showed that cdtR sequences from the four tcdC allele A isolates should have a predicted truncated protein sequence of 107 amino acids instead of 249, due to the presence of a stop codon TAA at positions 322 to 324 in place of a codon GAA. This mutation which might affect the functionality of the positive regulator of CDT production was retrieved from animal and human isolates. The third group was represented in the tcdC preliminary study by six isolates harboring a 54-bp deletion in tcdC. MLST studies (24, 25) and comparisons of whole genomes by microarray analyses (48) confirmed the clonality of A− B+ C. difficile toxin variants, as well as PCR ribotype 027 isolates. Triple-locus analysis and composite sequence analysis confirmed that PCR ribotype 027 isolates constituted a well-defined group. But analyses of genes in our study (Fig. 2 and 3) showed that PCR ribotype 027 isolates shared their tcdR allele (A) with isolates of some tcdC alleles (E to F and C) and their cdtR allele (A) with isolates of tcdC alleles C and J. In this study, evolutions of the three toxin regulatory genes tcdR and tcdC (the positive regulator and negative regulator of the production of large clostridial toxins TcdA and TcdB, respectively) and cdtR (the positive regulator of the production of binary toxin CDT) were found to be congruent in A− B+ toxin variants and isolates with a 39- or 54-bp deletion in tcdC. Among isolates possessing an 18-bp deletion in tcdC (associated or not with a 1-bp deletion at position 117), a 36-bp deletion, or no deletion (wt tcdC), evolutions of the three genes were more confused. These isolates were well separated in tcdC (alleles C, I, and J and E to F), intermixed in tcdR alleles A and B, and partly resolved in cdtR alleles A and B encompassing all isolates with an 18-bp deletion and a 36-bp deletion and in cdtR allele C encompassing all isolates with a wt tcdC gene.
The putative divergent evolution of tcdC and tcdR was rather surprising, since both genes are located within the same PaLoc and were both implicated in the regulation of tcdA and tcdB toxin production. tcdR had a low G+C% content (20.3 mol%) (this study and reference 24) compared to those of the other genes of the PaLoc, tcdA and tcdB (both 29 mol%) (24) and tcdC (32 mol%, this study), or the mean G+C% of C. difficile, ca. 29 mol% (strain 630). It has been suggested that TcdR and other clostridial sigma factors, BotR, TetR, and UviA, are all likely to be derived from the same ancestral protein whose original source is still unknown (13).
tcdC allele A isolates (39-bp deletion in tcdC, ST1, toxinotype V, binary toxin CDT positive, and assigned to PCR ribotype 078) represent a new homogeneous group of strains. Similar isolates have been circulating worldwide for several years. In Italy, Spigaglia and Mastrantonio (46) identified eight isolates of tcdC type A between 1991 and 1999 and seven isolates between 2000 and 2001. All isolates had the same PCR ribotype profile and were toxinotype V. In another study of 435 isolates collected in an Italian hospital during the period from 2000 to 2006, 22.8% of isolates were binary toxin CDT positive, and 80% of the CDT-positive isolates were toxinotype V, with the same PCR ribotype profile (M. P. Buttrini, P. Spigaglia, P. Somenzi, L. Zerbini, G. Dettori, C. Cheezi, P. Mastrantonio, and M. G. Menozzi, presented at the 2nd International C. difficile Symposium, Maribor, Slovenia, 2007). The authors indicate that a “variant clone” appeared at the end of 2000 in that hospital, with a peak in 2002 to 2003, and is still circulating. This clone with a 39-bp deletion in tcdC is often isolated from food animals (19, 44) and is currently suspected of being responsible for infection in humans (15, 38). From an extensive study of 650 recently obtained C. difficile isolates (from 2001 to 2006) and more than 6,000 historic isolates (before 2001), the authors identified 14 toxinotype V isolates from humans (7 isolates from 1990 to 2001 and 7 isolates from 2001 to 2006 [15]). They compared the human toxinotype V isolates with toxinotype V isolates from food animals. All human and animal isolates contained both the binary toxin and the 39-bp deletion in tcdC and belonged to restriction endonuclease analysis group BK and were PCR ribotype 078. One human-animal isolate pair was indistinguishable by pulsed-field gel electrophoresis and by restriction endonuclease analysis. Another work done in the United States identified C. difficile in 44.7% of 85 meat product samples, where 68.8% of isolates were PCR ribotype 078 (J. G. Songer, H. T. Trinh, A. D. Thompson, G. Killgore, L. C. McDonald, and B. M. Limbago, presented at the 2nd International C. difficile Symposium, Maribor, Slovenia, 2007). In a prospective study of C. difficile infections in Europe, Barbut et al. (2) have found among toxin-variant strains three prevalent toxinotypes, toxinotype III (n = 25), V (n = 28), and VIII (n = 22). Toxinotype V isolates might correspond to the new clone with a 39-bp deletion and a 078 PCR ribotype. In this study, toxinotype VIII isolates correspond to the A− B+ toxin variants and were PCR ribotype 017. In France, recent data obtained from the National Reference Centre network for C. difficile surveillance showed that the prevalence of isolates with a 39-bp deletion from human origin accounted for 10.5% of the total. This group of strains readily isolated from various food animals and retail meats and isolated from severe cases and clusters of cases of human infections in hospitals and communities might represent a new threat to human health and food safety. Further studies are needed to better understand their epidemiology and the possible role of food animals in the transmission to humans.
Acknowledgments
We thank French private and hospital laboratories for their participation in the French surveillance network for C. difficile.
We thank Frédéric Barbut for kindly providing some C. difficile strains harboring a 54-bp deletion in tcdC. Maryse Gibert is warmly thanked for performing the cytotoxicity experiments. We are indebted to Jean-Philippe Carlier for helpful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 1 October 2008.
REFERENCES
- 1.Barbut, F., D. Decre, V. Lalande, B. Burghoffer, L. Noussair, A. Gigandon, F. Espinasse, L. Raskine, J. Robert, A. Mangeol, C. Branger, and J. C. Petit. 2005. Clinical features of Clostridium difficile-associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J. Med. Microbiol. 54181-185. [DOI] [PubMed] [Google Scholar]
- 2.Barbut, F., P. Mastrantonio, M. Delmée, J. Brazier, E. Kuijper, and I. Poxton. 2007. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin. Microbiol. Infect. 131048-1057. [DOI] [PubMed] [Google Scholar]
- 3.Barbut, F., and J. C. Petit. 2001. Epidemiology of Clostridium difficile-associated infections. Clin. Microbiol. Infect. 7405-410. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. G. 2006. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann. Intern. Med. 145758-764. [DOI] [PubMed] [Google Scholar]
- 5.Baverud, V. 2002. Clostridium difficile infections in animals with special reference to the horse. A review. Vet. Q. 24203-219. [DOI] [PubMed] [Google Scholar]
- 6.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi, X., and L. F. Liu. 1996. A replicational model for DNA recombination between direct repeats. J. Mol. Biol. 256849-858. [DOI] [PubMed] [Google Scholar]
- 8.Carter, G. P., D. Lyras, D. L. Allen, K. E. Mackin, P. M. Howarth, J. R. O'Connor, and J. I. Rood. 2007. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J. Bacteriol. 1897290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curry, S. R., J. W. Marsh, C. A. Muto, M. M. O'Leary, A. W. Pasculle, and L. H. Harrison. 2007. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J. Clin. Microbiol. 45215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmée, M., M. Homel, and G. Wauters. 1985. Serogrouping of Clostridium difficile strains by slide agglutination. J. Clin. Microbiol. 21323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmée, M., Y. Laroche, V. Avesani, and G. Cornelis. 1986. Comparison of serogrouping and polyacrylamide gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 24991-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66206-219. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy, B., and S. Matamouros. 2006. Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors. Res. Microbiol. 157201-205. [DOI] [PubMed] [Google Scholar]
- 14.Hundsberger, T., V. Braun, M. Weidmann, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244735-742. [DOI] [PubMed] [Google Scholar]
- 15.Jhung, M. A., A. D. Thompson, G. E. Killgore, W. E. Zukowski, G. Songer, M. Warny, S. Johnson, D. N. Gerding, L. C. McDonald, and B. M. Limbago. 2008. Toxinotype V Clostridium difficile in humans and food animals. Emerg. Infect. Dis. 141039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, S., S. P. Sambol, J. S. Brazier, M. Delmée, V. Avesani, M. M. Merrigan, and D. N. Gerding. 2003. International typing study of toxin A-negative, toxin B-positive Clostridium difficile variants. J. Clin. Microbiol. 411543-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 171230-1231. [DOI] [PubMed] [Google Scholar]
- 18.Joseph, R., D. Demeyer, D. Vanrenterghem, R. van den Berg, E. Kuijper, and M. Delmee. 2005. First isolation of Clostridium difficile PCR ribotype 027, toxinotype III in Belgium. Euro. Surveill. 10E051020.4. [DOI] [PubMed] [Google Scholar]
- 19.Keel, M. K., J. S. Brazier, K. W. Post, S. Weese, and J. G. Songer. 2007. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves and other species. J. Clin. Microbiol. 451963-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16111-120. [DOI] [PubMed] [Google Scholar]
- 21.Kuijper, E. J., B. Coignard, and P. Tull. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin. Microbiol. Infect. 12(Suppl. 6)2-18. [DOI] [PubMed] [Google Scholar]
- 22.Kuijper, E. J., R. J. van den Berg, S. Debast, C. E. Visser, D. Veenendaal, A. Troelstra, T. van der Kooi, S. van den Hof, and D. W. Notermans. 2006. Clostridium difficile ribotype 027, toxinotype III, the Netherlands. Emerg Infect. Dis. 12827-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 24.Lemée, L., I. Bourgeois, E. Ruffin, A. Collignon, J. F. Lemeland, and J. L. Pons. 2005. Multilocus sequence analysis and comparative evolution of virulence-associated genes and housekeeping genes of Clostridium difficile. Microbiology 1513171-3180. [DOI] [PubMed] [Google Scholar]
- 25.Lemée, L., A. Dhalluin, M. Pestel-Caron, J. F. Lemeland, and J. L. Pons. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 422609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 3532442-2449. [DOI] [PubMed] [Google Scholar]
- 27.Louther, J. 1996. Enteric precautions for Clostridium difficile. Am. J. Nurs. 9619. [DOI] [PubMed] [Google Scholar]
- 28.Lyerly, D. M., H. C. Krivan, and T. D. Wilkins. 1988. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacCannell, D. R., T. J. Louie, D. B. Gregson, M. Laverdiere, A. C. Labbe, F. Laing, and S. Henwick. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 442147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 953140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani, N., and B. Dupuy. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. USA 985844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matamouros, S., P. England, and B. Dupuy. 2007. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 641274-1288. [DOI] [PubMed] [Google Scholar]
- 33.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 3532433-2441. [DOI] [PubMed] [Google Scholar]
- 34.Pépin, J., L. Valiquette, M. E. Alary, P. Villemure, A. Pelletier, K. Forget, K. Pepin, and D. Chouinard. 2004. Clostridium difficile-associated diarrhea in a region of Québec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perelle, S., M. Gibert, P. Bourlioux, G. Corthier, and M. R. Popoff. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect. Immun. 651402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popoff, M. R., E. J. Rubin, D. M. Gill, and P. Boquet. 1988. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 562299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Palacios, A., H. R. Stampfli, T. Duffield, A. S. Peregrine, L. A. Trotz-Williams, L. G. Arroyo, J. S. Brazier, and J. S. Weese. 2006. Clostridium difficile PCR ribotypes in calves, Canada. Emerg. Infect. Dis. 121730-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rupnik, M. 2007. Is Clostridium difficile-associated infection a potentially zoonotic and foodborne disease? Clin. Microbiol. Infect. 13457-459. [DOI] [PubMed] [Google Scholar]
- 39.Rupnik, M., B. Dupuy, N. F. Fairweather, D. N. Gerding, S. Johnson, I. Just, D. M. Lyerly, M. R. Popoff, J. I. Rood, A. L. Sonenshein, M. Thelestam, B. W. Wren, T. D. Wilkins, and C. von Eichel-Streiber. 2005. Revised nomenclature of Clostridium difficile toxins and associated genes. J. Med. Microbiol. 54113-117. [DOI] [PubMed] [Google Scholar]
- 40.Rupnik, M., M. Grabnar, and B. Geric. 2003. Binary toxin producing Clostridium difficile strains. Anaerobe 9289-294. [DOI] [PubMed] [Google Scholar]
- 41.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 42.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38779-786. [DOI] [PubMed] [Google Scholar]
- 43.Songer, J. G. 2004. The emergence of Clostridium difficile as a pathogen of food animals. Anim. Health Res. Rev. 5321-326. [DOI] [PubMed] [Google Scholar]
- 44.Songer, J. G., and M. A. Anderson. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 121-4. [DOI] [PubMed] [Google Scholar]
- 45.Songer, J. G., and F. A. Uzal. 2005. Clostridial enteric infections in pigs. J. Vet. Diagn. Investig. 17528-536. [DOI] [PubMed] [Google Scholar]
- 46.Spigaglia, P., and P. Mastrantonio. 2004. Comparative analysis of Clostridium difficile clinical isolates belonging to different genetic lineages and time periods. J. Med. Microbiol. 531129-1136. [DOI] [PubMed] [Google Scholar]
- 47.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 403470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. S. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 1887297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stare, B. G., M. Delmee, and M. Rupnik. 2007. Variant forms of the binary toxin CDT locus and tcdC gene in Clostridium difficile strains. J. Med. Microbiol. 56329-335. [DOI] [PubMed] [Google Scholar]
- 50.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186307-312. [DOI] [PubMed] [Google Scholar]
- 51.Tachon, M., C. Cattoen, K. Blanckaert, I. Poujol, A. Carbonne, F. Barbut, J. C. Petit, and B. Coignard. 2006. First cluster of C. difficile toxinotype III, PCR-ribotype 027 associated disease in France: preliminary report. Euro. Surveill. 11E060504.1. [DOI] [PubMed] [Google Scholar]
- 52.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 3661079-1084. [DOI] [PubMed] [Google Scholar]