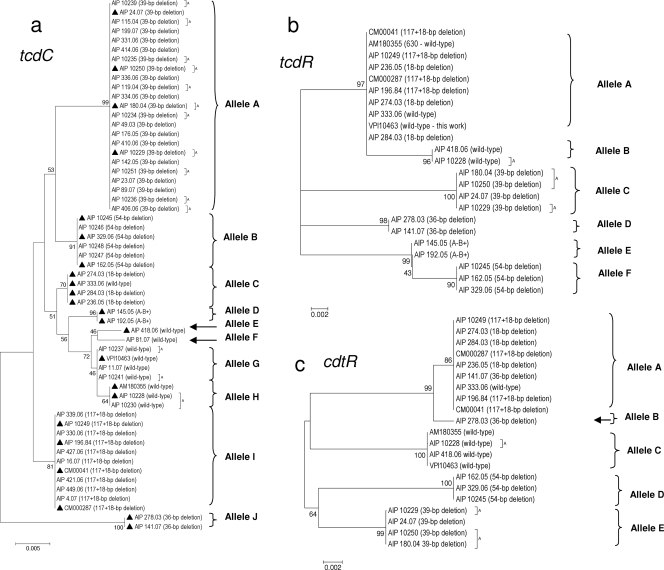
FIG. 1.
Dendrograms showing genetic relationships of the C. difficile isolates based on allele sequences of individual toxin regulatory loci tcdC (a), tcdR (b), and cdtR (c). Fifty-seven isolates are shown for tcdC. From these 57 isolates, 23 isolates were selected (▴) for tcdR and cdtR analysis. Twenty-one isolates are shown for the cdtR locus (lack of amplification for the two A− B+ isolates). Dendrograms were reconstructed from the nucleotide sequence of each gene by using the neighbor-joining method. The genetic distances were computed by using the Kimura two-parameter model. The scale bar indicates the genetic distance. The number shown next to each node indicates the percent bootstrap value of 1,000 replicates. A, animal isolate.