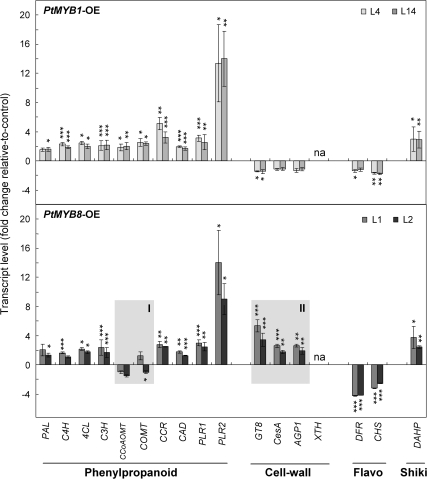
Fig. 4.
Targeted RT-qPCR analysis in PtMYB1 and PtMYB8 transgenic plantlets: validation of microarray and expression data from genes related to secondary metabolism and cell wall assembly. Phenylpropanoid-related genes are phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), coumarate 3-hydroxylase (C3H), caffeate O-methyltransferase (COMT), caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), cinnamyl-alcohol dehydrogenase (CAD), and pinoresinol-lariciresinol reductase 1 and 2 (PLR1 and PLR2); cell wall-related genes are cellulose synthase (CesA), glycosyltransferase family 8 (GT8), arabinogalactan (AGP1), and xyloglucan endotransglycosylase/hydrolase (XTH). Flavonoid related genes (Flavo) are chalcone synthase (CHS), dihydroflavonol 4-reductase (DFR); the shikimate-related gene (Shiki) is 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetase (DAHP). For both transgenics, transcript accumulation was assessed by RT-qPCR on 3-week-old plantlets produced from two independent transgenic lines for each gene construct (L4 and L14 for PtMYB1; L1 and L2 for PtMYB8). Transcript levels were determined from four biological replicates (25 plantlets per replicate) and were normalized relative to EF1-alpha expression level. Transcript levels were then expressed relative to control plants, and the significance of differential transcript accumulation was evaluated with Student's t-test (two-sample, unpaired, one-sided) at P ≤0.05 (*), P ≤0.01 (**), and P ≤0.001 (***). na, Not amplified in MYB transgenics.