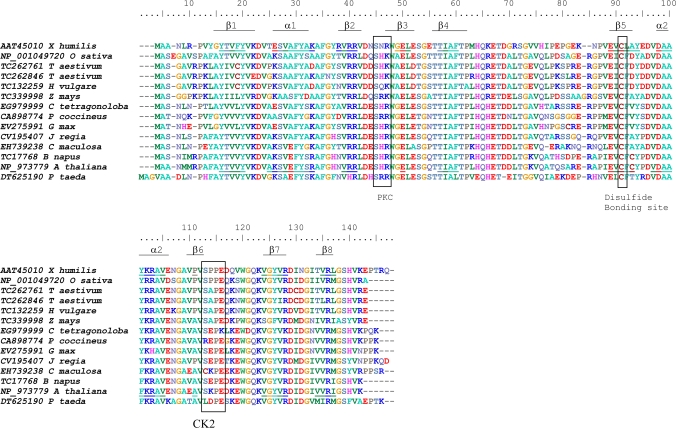
Fig. 2.
ClustalW alignment of predicted amino acid sequences of plant dsi-1VOC orthologues identified in the Genbank and TIGR EST databases, including X. humilis, rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), maize (Zea mays), guar (Cyamopsis tetragonoloba), runner bean (Phaseolus coccineus), soybean (Glycine max), walnut (Juglans regia), spotted knapweed (Centaurea maculosa), oilseed rape (Brassica napus), Arabidopsis thaliana, and Loblolly pine (Pinus taeda). Boxed amino acids demarcate conserved phosphorylation sites for protein kinase C (PKC) and protein casein kinase II (CK2) identified by PROSITE, and a potential disulphide bonding site predicted by DISULFIND. Amino acids in the X. humilis and A. thaliana dsi-1VOC orthologues which are predicted to fold into the βαβββ structural repeats characteristic of the VOC superfamily, are underlined.