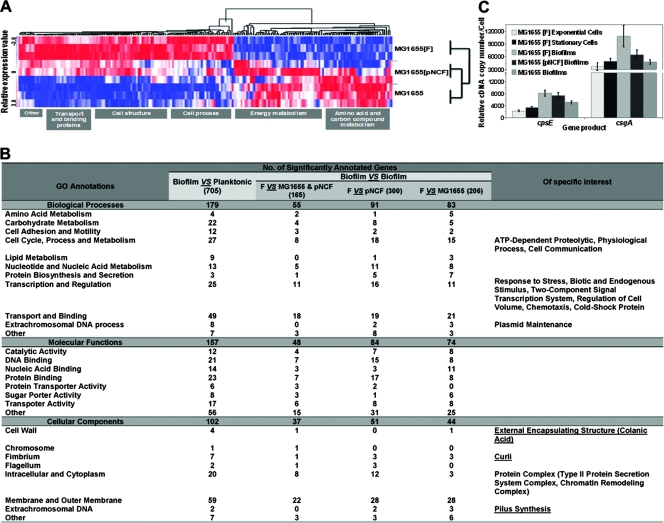
FIG. 4.
Genes regulating F-pilus, colanic acid, and curli formation are differentially expressed in biofilms containing a natural F plasmid. (A) Dynamic hierarchical clustering of E. coli gene expression during biofilm maturation at 48 h. Each row shows data for one culture form. Each column shows data for one gene. Red, upregulated; white, no change; blue, downregulated. The sample profiles were grouped into two clusters: MG1655(F) versus MG1655 (plasmid-free) and MG1655(pNCF) biofilms. (B) GO annotations overrepresented in an F-plasmid-containing biofilm compared with the other conditions tested. Note that an Affymetrix E. coli antisense GeneChip consists of 7,312 probe sets. These probe sets are placed into three ontology branches, the biological processes (2,979 probe sets), the molecular functions (3,228 probe sets), and the cellular components (1,999 probe sets), according to the NetAffx Analysis Center. The significant GO terms in this study are shown. The numbers of significantly differentially expressed genes are indicated. The last column indicates the specific GO characteristics of the significantly differentially expressed genes mediated by a natural conjugative F plasmid. (C) csgA and cpsE expression levels determined by quantitative RT-PCR. Note the increases in the csgA and cpsE expression levels in MG1655(F) and MG1655(pNCF) biofilms (P ≤ 0.05). All experiments were repeated three times, and the average levels of expression and standard derivations are indicated.