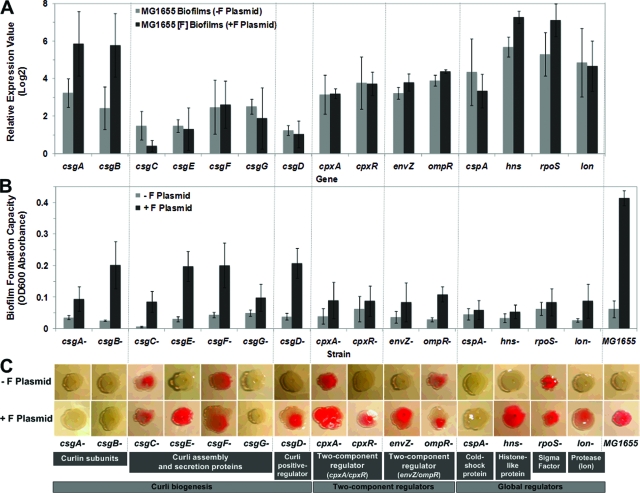
FIG. 5.
Natural F plasmid helped E. coli produce curli during biofilm maturation. (A) Levels of expression of 15 curli biogenesis- and regulation-related genes obtained from microarray analysis. The bars indicate the averages for three replicates, and the error bars indicate the standard deviations; the data are representative of the data from two independent experiments. Asterisk, P < 0.05 compared with a plasmid-free biofilm. (B) Crystal violet biofilm formation assay showing that most curli biogenesis- and regulation-related mutants were likely dependent on the presence of F plasmid during biofilm formation, but the csgG, cpxA, cpxR, cspA, hns, rpoS, and lon mutants were less dependent (P > 0.05). The bars indicate the averages for eight replicates, and the error bars indicate the standard deviations; the data are representative of the data from 10 independent experiments. Asterisk, P < 0.05 compared with a plasmid-free biofilm. OD600, optical density at 600 nm. (C) Congo red assay showing that curli production was induced by introducing F plasmid only into cpxR, csgD, csgE, envZ, hns, and lon mutants at 37°C. Assays were repeated 10 times, and representative images of Congo red binding are shown.