Abstract
A bacterium, strain BC, was isolated from a benzene-degrading chlorate-reducing enrichment culture. Strain BC degrades benzene in conjunction with chlorate reduction. Cells of strain BC are short rods that are 0.6 μm wide and 1 to 2 μm long, are motile, and stain gram negative. Strain BC grows on benzene and some other aromatic compounds with oxygen or in the absence of oxygen with chlorate as the electron acceptor. Strain BC is a denitrifying bacterium, but it is not able to grow on benzene with nitrate. The closest cultured relative is Alicycliphilus denitrificans type strain K601, a cyclohexanol-degrading nitrate-reducing betaproteobacterium. Chlorate reductase (0.4 U/mg protein) and chlorite dismutase (5.7 U/mg protein) activities in cell extracts of strain BC were determined. Gene sequences encoding a known chlorite dismutase (cld) were not detected in strain BC by using the PCR primers described in previous studies. As physiological and biochemical data indicated that there was oxygenation of benzene during growth with chlorate, a strategy was developed to detect genes encoding monooxygenase and dioxygenase enzymes potentially involved in benzene degradation in strain BC. Using primer sets designed to amplify members of distinct evolutionary branches in the catabolic families involved in benzene biodegradation, two oxygenase genes putatively encoding the enzymes performing the initial successive monooxygenations (BC-BMOa) and the cleavage of catechol (BC-C23O) were detected. Our findings suggest that oxygen formed by dismutation of chlorite can be used to attack organic molecules by means of oxygenases, as exemplified with benzene. Thus, aerobic pathways can be employed under conditions in which no external oxygen is supplied.
Contamination of the groundwater with perchlorate (ClO4−) and chlorate (ClO3−) has been observed at many places, especially in the United States (50). The presence of perchlorate in the environment has been associated primarily with manufacturing, handling, and dismantling of munitions, since it is used as a major component of rocket propellants and explosives. (Per)chlorate contamination poses a significant health threat, as toxicological studies have demonstrated (1). Research during the last decade has resulted in isolation of various (per)chlorate-reducing bacteria and characterization of the key enzymes in these isolates. Perchlorate and chlorate are ideal electron acceptors for microorganisms due to their high redox potentials (ClO4−/Cl− E0, 1.287 V; ClO3−/Cl− E0, 1.03 V) (12). (Per)chlorate-reducing microorganisms reduce (per)chlorate to chloride. Typically, during (per)chlorate reduction the intermediate chlorite is dismutated to molecular oxygen and chloride (28, 37, 52). This means that (per)chlorate-reducing microorganisms can produce oxygen having a metabolic origin in anaerobic environments. The formation of oxygen during (per)chlorate reduction may result in rapid oxidation of compounds which are slowly degraded under anaerobic conditions (13).
An example of such a compound is benzene. Generally, benzene is rapidly degraded by aerobic microorganisms with the appropriate catabolic potential, but in anaerobic environments biodegradation is much slower (30). Anaerobic biodegradation of benzene under various redox conditions has been described, but only in a few studies were the microorganisms involved identified (10, 23, 35, 39, 49). So far, only four anaerobic benzene-degrading bacteria have been described, two Dechloromonas strains (strains RCB and JJ) that degrade benzene in conjunction with (per)chlorate (only strain RCB), nitrate, or oxygen reduction (9) and two denitrifying Azoarcus strains (strains DN11 and AN9) (23). The optimal physiological conditions for anaerobic benzene-degrading bacteria and the biodegradation pathways are still largely unclear (6, 8, 17, 26, 33, 48).
The relatively high solubility of benzene, toluene, ethylbenzene, and xylene (the so-called BTEX compounds) and the low solubility of oxygen often result in BTEX contamination in anoxic zones of the environment. Anaerobic bioremediation is attractive when anaerobic conditions prevail at a polluted soil site. The potential for using (per)chlorate-reducing microorganisms for bioremediation of soils and sediments has been recognized in previous studies (13, 29). It has been demonstrated that addition of (per)chlorate-reducing microorganisms and chlorite to an anoxic soil led to complete degradation of [14C]benzene to 14CO2 (13). Toluene degradation was observed in sand columns inoculated with toluene-degrading and chlorate-reducing enrichment cultures (29). In another study, addition of chlorate to a soil column polluted with benzene resulted in removal of benzene in conjunction with chlorate reduction (45). Recently, we obtained a highly active benzene-degrading, chlorate-reducing enrichment culture with mixed material from a wastewater treatment plant and soil samples, and we analyzed which phylogenetic groups of bacteria were present (54). Here, we describe isolation of strain BC from this enrichment culture. This bacterium is capable of growth on benzene with chlorate as the electron acceptor. Our data suggest that oxygen produced in the dismutation of chlorite is used to degrade benzene, and oxygenase systems potentially involved in this process were identified.
MATERIALS AND METHODS
Inoculum and cultivation and isolation procedures.
Strain BC was isolated from a stable benzene-degrading, chlorate-reducing enrichment culture. Isolation and cultivation of strain BC were performed using strictly anaerobic AW-1-sulfate medium (54). Cultures were incubated at 30°C in the dark in an orbital shaker (50 rpm) in 120-ml serum bottles containing 40 ml medium. Soluble electron donors and acceptors were added from sterile, anaerobic stock solutions. Oxygen was added to the gas phase from a sterile 100% oxygen gas stock. Benzene was added from a water-saturated stock solution (20 mM) and was readded to batches when it was depleted. Chlorate was added from a 0.4 M NaClO3 stock solution. Where indicated below, fermented yeast extract (FYE) (0.125 g/liter) was added to the enrichment as a nutrient supplement to stimulate growth (21, 51, 54).
For strain isolation, dilution series of the stable enrichment culture were prepared using AW-1-sulfate medium with benzene (0.25 mM) and chlorate (10 mM) as energy substrates, 0.125 g/liter FYE as a nutrient supplement, and 1.2% agar (Noble agar; Difco, Becton Dickinson Microbiology Systems, Sparks, MD) to solidify the medium. Colonies were picked from the highest dilutions and transferred to new agar dilution series. This procedure was repeated four times. The purity was checked by microscopic observation of cultures grown on benzene and easily degradable substrates (e.g., yeast extract plus acetate). Furthermore, denaturing gradient gel electrophoresis (DGGE) was used to confirm the purity of the cultures. Strain BC was routinely grown with benzene (0.25 or 0.5 mM) and chlorate (10 mM).
The Gram type was determined using Gram staining and electron microscopy as previously described (36). Phase-contrast micrographs were obtained with a Leica (Wetzlar, Germany) DMR HC microscope equipped with a Leica DC 250 digital camera. The Leica QWin computer program was used to obtain digital micrographs.
Physiological studies.
All growth parameters of strain BC were determined using either duplicate or triplicate batches of AW-1-sulfate medium (without FYE). When necessary, all electron donors and electron acceptors were added as sodium salts. The growth rate of strain BC was determined by determining the increase in the optical density at 600 nm (OD600) and/or the increase in the number of cells with time for triplicate batches. Numbers of cells were determined by phase-contrast microscopy using a Bürker-Türk counting chamber at a magnification of ×1,000. Cell yields were determined by determining the dry weight of the biomass (in 200-ml cultures). The dry weight was determined gravimetrically after the cell pellet was dried at 105°C overnight. The optimum pH was determined with acetate (10 mM) and nitrate (10 mM) at 30°C using a pH range from 6.6 to 9.0 for duplicate batches. Different pH values for the medium were obtained by changing the percentage of CO2 in the headspace while the bicarbonate concentration in the medium was kept constant, as described previously (59). The optimum temperature was determined with acetate (10 mM) and nitrate (10 mM) using temperatures ranging from 4 to 55°C and duplicate batches.
To determine the substrate spectrum of strain BC, the following electron donors (at a concentration of 10 mM unless indicated otherwise) were tested using duplicate batches with nitrate (10 mM) as the electron acceptor: acetate, lactate, pyruvate, succinate, propionate, butyrate, malate, citrate, fumarate, glucose, fructose, xylose, alanine, glycine, glutamate, ethanol, methanol, glycerol, Fe(II)Cl2 (5 mM), Na2S (1 mM), H2 (170 kPa, with 1 mM acetate added), and yeast extract (1 g/liter). Late-log-phase cells of strain BC grown on acetate (10 mM) and nitrate (10 mM) were used as the inoculum (5%) in this experiment. The following electron donors were tested using duplicate batches with either nitrate (10 mM) or oxygen (5% in the headspace): benzene (0.25 mM), toluene (0.25 mM), ethylbenzene (0.10 mM), o-xylene (0.10 mM), m-xylene (0.10 mM), p-xylene (0.10 mM), monochlorobenzene (0.05 mM), benzoate (1 mM), phenol (1 mM), cyclohexanol (1 mM), p-hydroxybenzoate (1 mM), o-cresol (1 mM), m-cresol (1 mM), p-cresol (1 mM), and catechol (1 mM). Late-log-phase cells of strain BC grown on either acetate (10 mM) and nitrate (10 mM) or benzene (0.5 mM added repeatedly) and oxygen (5% in the headspace) were used as the inoculum (5%) in this experiment. Furthermore, in addition to benzene the following electron donors were tested with chlorate (10 mM) as the electron acceptor: acetate (10 mM), toluene (0.25 mM), phenol (1 mM), o-cresol (1 mM), m-cresol (1 mM), p-cresol (1 mM), and catechol (1 mM). Late-log-phase cells of strain BC grown on benzene (0.5 mM added repeatedly) and chlorate (10 mM) were used as the inoculum (5%) in this experiment. Growth was monitored by visual observation of turbidity and by determining the decrease in the nitrate or chlorate concentration.
The following electron acceptors (at a concentration of 10 mM unless indicated otherwise) were tested using duplicate batches with acetate (10 mM) as the electron donor: oxygen (10% in the headspace), perchlorate, chlorate, nitrate, nitrite (5 mM), sulfate, sulfite, thiosulfate, fumarate, manganese(IV) oxide (20 mmol/liter), iron(III) pyrophosphate, iron(III) nitrilotriacetic acid, anthraquinone-2,6-disulfonate (4 mM), bromate (5 and 10 mM), selenate (5 and 10 mM), and arsenate (5 and 10 mM). Electron acceptor use was monitored by visual observation and by measuring the acetate concentration and also the decrease in the level of the electron acceptor when the acetate concentration decreased.
Analytical procedures.
Benzene contents were measured by headspace analysis using a gas chromatograph as described previously (54). Anion (chlorate, chloride, and nitrate) contents were determined by high-pressure liquid chromatography as described previously (42). Oxygen in the headspace of batches was measured with a gas chromatograph as described previously (44). Catechol, benzoate, and phenol were analyzed by using a high-pressure liquid chromatograph equipped with a Chrompack column and Chromspher 5 pesticides (100 by 3 mm). The mobile phase consisted of different ratios of 0.1% trifluoroacetic acid and 50% acetonitrile plus 50% trifluoroacetic acid (0.1%) at a flow rate of 0.6 ml/min. The detector was a Spectra System UV1000 detector.
Molecular biological techniques.
A bead-beating and phenol-chloroform-based DNA extraction method was used to extract DNA from pure cultures of strain BC (51). Amplification with primers 7f and 1492 and purification and sequencing of the 16S rRNA genes were performed as previously described (51). Part of the 16S rRNA gene (424 bp) of strain BC was analyzed, and its sequence exhibited 100% similarity with the 16S rRNA gene sequence of clone cA8 (1,487 bp) obtained from the enrichment (54). In sequence analysis studies, the 1,487-bp 16S rRNA gene sequence fragment was used. Sequences of 16S rRNA genes were compared with sequences deposited in publicly accessible databases using the NCBI BLAST search tool at http://www.ncbi.nlm.nih.gov/blast/ (2, 31). DNA from strain BC was used as the PCR template for DGGE as described previously (54). Silver staining and development of the gels were performed by using the method of Sanguinetti et al. (41). A phylogenetic tree of partial 16S rRNA gene sequences of strain BC and related bacteria was constructed. Alignment and phylogenetic analysis were performed with the ARB software, and the tree was constructed using the neighbor-joining method based on Escherichia coli positions 101 to 1221 and a 50% conservation filter for Betaproteobacteria, as implemented in ARB. The accession numbers of strain BC and Alicycliphilus denitrificans type strain K601 in the GenBank database are DQ342277 and AJ418042, respectively. Strain BC has been deposited in two different collection of microorganisms, the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) as strain DSM 18852 and the Japanese Collection of Microorganisms (Riken BioResource Center, Japan) as strain JCM 14587.
To detect chlorite dismutase gene (cld) sequences, we used the conditions described elsewhere with primers UCD-238F and UCD-646R and primers DCD-F and DCD-R (4). Additionally, we tested modified annealing temperatures using a range of ±10°C. DNA from Pseudomonas chloritidismutans isolated in our laboratory was used as a positive control (59).
To detect the benzene and catechol oxygenase genes present in strain BC, we designed the following primer sets targeting three different evolutionary clusters around the I.2.A and I.2.B sequence spaces of the type I extradiol dioxygenase (EXDO) family (16) in an upgraded (2006) database of related sequences (all primer sequences are 5′-3′ sequences): forward primer EXDO-A-F (ATG AAV AAA GGH GTW HTG CGH CCN GG) and reverse primer EXDO-A-R1 (GYG GCC ADG CYT CNG GRT TG) (expected product size, ∼430 bp) or EXDO-A-R2 (ATR TCV AKV GAD GTR TCG STC ATG) (expected product size, ∼730 bp); forward primer EXDO-B-F (TRA CMG GHG TNH TGC GYC CVG GSC A) and reverse primer EXDO-B-R (GCC RTG VCG SGT BGG VCC GAT) (expected PCR fragment size, ∼750 bp); forward primer EXDO-C-F (CAY TAY CGY GAC CGK ATY GG) and reverse primer EXDO-C-R1 (TCR TCA TGB GCY TTR TTG CTG CA) (expected product size, ∼530 bp) or EXDO-C-R2 (TCG TTS CGR TTD CCS GAV GGR TCG AAG AA) (expected product size, ∼710 bp).
To detect genes encoding the large subunit of the four-component alkene/aromatic monooxygenase and phenol hydroxylase/toluene monooxygenase (ring-hydroxylating monooxygenase [RHMO]) members (RHMO-TMOPHE) of the soluble diiron monooxygenase family (27), the following primers were designed: forward primer RHMO-TMOPHE-F (GAY CCB TTY CGY HTR ACC ATG GA) and reverse primer RHMO-TMOPHE-R (GGC ARC ATG TAR TCC WKC ATC AT). The expected amplification product size was ∼701 bp. For amplification of four-component aromatic monooxygenase large subunits, mainly comprising toluene/benzene monooxygenases (RHMO-T/BMO), the primers used were forward primer RHMO-T/BMO-F (ASR AAC TGC ATR TTG GTR AAR CC) and reverse primer RHMO-T/BMO-R (GAR TAC GTS MGB RTY CAR CGX GAR AAG GA), which annealed from position 169 to position 617 in the Pseudomonas mendocina KR1 tmoA coding DNA sequence and produced an expected PCR product that was 448 bp long.
Common PCR conditions used for amplification screening for the presence of the oxygenase genes targeted (described above) were as follows (final volume of the PCR mixture, 50 μl): 1× colorless GoTaq reaction buffer (Promega, Madison, WI), 5 U of GoTaq polymerase (Promega, Madison, WI), each deoxynucleoside triphosphate (Fermentas) at a concentration of 200 μM, and 10 pmol of each primer (synthesized by Invitrogen GmbH, Karlsruhe, Germany). For thermal cycling, an Eppendorf gradient themocycler was used as follows: one initial denaturation step at 94°C for 1 min, followed by 35 cycles consisting of 45 s at 94°C, 45 s at 50, 55, or 60°C, and 1 min at 72°C and then one final elongation step at 72°C for 7 min. Polymerization reactions were stopped by cooling the samples at 4°C. Reactions were further analyzed by 1× Tris-acetate-EDTA-1% agarose gel electrophoresis to assess the presence of PCR products that were the expected sizes. For DNA sequencing, the PCR product was purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) when a single size was observed. When the PCR products were different sizes, the product size matching the expected fragment size was excised from the agarose gel and purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany), and this purified fragment was then subjected to direct sequencing. DNA sequencing was performed according to the manufacturer's instructions using 600 ng of the purified PCR products as DNA templates in two independent sequencing reactions with the same primers that were used for the original PCR amplification and a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA). The sequence fragments were detected with a 3130xl DNA capillary sequencer-genetic analyzer (Applied Biosystems, Foster City, CA). The corresponding sequence chromatogram reads were assembled using Sequencher software, version 4.0 (Genes Codes Corporation, Ann Arbor, MI), and the assembled contigs were exported as text files. The sequences were oriented in the same direction (5′-3′) relative to the coding DNA sequence.
For each sequence, the conceptual translation of a peptide in one frame not producing stop codons was confirmed using standard and bacterial translation codes with GeneDoc multiple-sequence alignment editor software (34). DNA sequences were used for BLAST searches with the tblastx option and the nonredundant database in order to confirm that they belonged to the gene family targeted (the highest scores were observed by comparison with the known family members expected). Additionally, for each sequence whether the conserved protein domain for the family targeted was maintained in the span analyzed was determined. The putative protein sequences obtained from strain BC were used to predict a model for the protein structure using the resolved crystal structure with a higher level of identity as the template and the (PS)2 modeling software (Protein Structure Prediction Server using RAMP model building) (11). These amino acid sequences were also later aligned with the translated multiple-sequence alignments for the corresponding families used for primer design using Clustal W and default values (47). A block of protein sequence alignments was selected with GeneDoc software (34). This block of sequences was aligned again, and neighbor-joining trees were constructed together with generation of bootstrap values for 1,000 trials using the functions implemented in Clustal W. For minor modifications in sequence alignments or neighbor-joining tree presentation, vectorial representations were imported into Openoffice suite 2.0.4 (OpenOffice) (http://www.openoffice.org/) from the alignment graphical view available from the BioEdit software (BioEdit) (http://www.mbio.ncsu.edu/) or from the graphical display of neighbor-joining tree files available for MEGA 3.1 software (25), selecting rooting on midpoint and arranging taxa for balanced shape.
Preparation of cell extracts and measurement of enzyme activity.
For preparation of cell extract, strain BC was grown in AW-1-sulfate medium (200 ml) with benzene and chlorate and with benzene and oxygen. Cell extracts were prepared as described by Wolterink et al. (59), except that centrifugation was performed at 13,000 rpm for 30 min at 4°C. Chlorate reductase and chlorite dismutase activities were determined using cell extracts of strain BC. The chlorate reductase activity was determined spectrophotometrically as described previously by monitoring the oxidation of reduced methyl viologen at 578 nm and 30°C (24). One unit of activity was defined as the amount of enzyme required to reduce 1 μmol of chlorate per min. The chlorite dismutase activity was determined by measuring the oxygen production with a Clark-type oxygen electrode (Yellow Spring Instruments, Yellow Springs, OH) as described previously (59)]. One unit of activity was defined as the amount of enzyme required to convert 1 μmol of chlorite per min. The protein content of the cell extract was determined by using the method of Bradford, with bovine serum albumin as the standard (7).
Nucleotide sequence accession numbers.
The DNA sequences obtained in this study have been deposited in the EMBL/GenBank/DBBJ databases under accession numbers EF596778 (670 bp) for the putative benzene monooxygenase large subunit (BC-BMOa)-encoding gene of Alicycliphilus sp. strain BC and EF596779 (707 bp) for the putative catechol 2,3-dioxygenase (BC-C23O)-encoding gene of Alicycliphilus sp. strain BC.
RESULTS
Isolation.
A stable enrichment culture that degraded benzene with chlorate as the electron acceptor was obtained previously (54). DGGE of 16S rRNA gene fragments followed by cloning and sequencing showed that at least four different bacterial species were numerically dominant in the culture. Initially, isolation of a benzene-degrading, chlorate-reducing bacterium from this enrichment culture was not successful. At a later stage, 0.125 g/liter FYE was added to the enrichment as a nutrient supplement to stimulate growth. This resulted in a decreased lag phase for benzene degradation (54). Subsequently, a pure culture of a benzene-degrading, chlorate-reducing bacterium, strain BC, was successfully isolated by using serial dilution techniques with agar (1.2%) and liquid media (59). Benzene (250 μM) and chlorate (10 mM) were added as the electron donor and acceptor, respectively, and FYE (0.125 g/liter) was added as a nutrient supplement. Within 3 to 4 weeks, colonies appeared in the agar dilutions. The colonies were round, lens shaped, 0.5 to 1.0 mm in diameter, and brown. Colonies were picked from the highest dilution with a sterile needle and directly transferred to new agar (and liquid) dilution series. This procedure was repeated until a pure culture was obtained. The purity of the culture was confirmed by microscopic observation and DGGE analysis of cultures grown on benzene and easily degradable substrates (e.g., yeast extract plus acetate). Identical morphologies and patterns were observed in these cultures by using microscopy and DGGE, respectively. The strain BC cells were short rods that were 0.6 μm wide and 1 to 2 μm long (Fig. 1), were motile, and stained gram negative. Flocculated growth of cells of strain BC was observed, especially during growth with chlorate as the electron acceptor and to a lesser extent during growth with nitrate or oxygen as the electron acceptor. Growth of strain BC occurred at a pH range from 6.6 to 9.0, and the optimum pH was 7.3. The optimal temperature for growth was 30 to 37°C, and the lower and upper limits for growth were about 15 and 40°C.
FIG. 1.
Phase-contrast micrograph of cells of strain BC grown on benzene and chlorate. Magnification, ×1,000.
Phylogenetic characterization: 16S rRNA gene sequence.
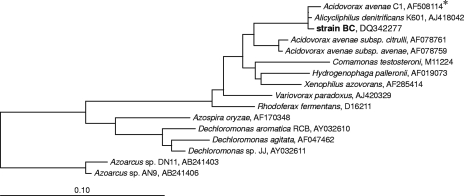
16S rRNA gene sequence analysis showed that strain BC is closely related to Acidovorax avenae isolate C1 and A. denitrificans type strain K601, and the levels of similarity were 99.9 and 99.7%, respectively (3, 32). Strain BC exhibited only 96% 16S rRNA gene sequence similarity with A. avenae subsp. avenae DSM 7227T (57). Therefore, we consider A. denitrificans K601T the closest cultured relative of strain BC, and in further physiological and (chemo)taxonomical characterization experiments A. denitrificans K601T was included as a reference strain. Some key physiological and (chemo)taxonomic characteristics of the two strains are described below.
Strain BC clusters in the family Comamonadaceae in the Betaproteobacteria (56) (Fig. 2). This family comprises several genera, including Acidovorax, Comamonas, Delftia, Hydrogenophaga, Rhodoferax, Brachymonas, Polaromonas, Variovorax, Xylophilus (55), and Xenophilus (5). Members of this phenotypically heterogeneous family are phylogenetically closely related to strain BC. Species belonging to the genera Dechloromonas and Azospira (formerly Dechlorosoma) in the beta subclass of the Proteobacteria were also included in the phylogenetic tree (Fig. 2), because (per)chlorate-reducing bacteria in the environment are predominantly members of the genera Dechloromonas and Azospira. The Dechloromonas strains capable of degrading benzene anaerobically, Dechloromonas sp. strains RCB and JJ, were also included in the phylogenetic tree. Both of these strains exhibited 91% similarity with strain BC based on 16S rRNA gene sequences, while Azoarcus sp. strains AN9 and DN11 capable of degrading benzene with nitrate were more distantly related and exhibited only 88% (AN9) and 87% (DN11) 16S rRNA gene sequence similarity with strain BC, respectively.
FIG. 2.
Phylogenetic tree of partial 16S rRNA gene sequences, showing the relationships between strain BC and some other members of the family Comamonadaceae in the Betaproteobacteria. Alignment and phylogenetic analysis were performed with the ARB software, and the tree was constructed using the neighbor-joining method based on E. coli positions 101 to 1221 and a 50% conservation filter for Betaproteobacteria, as implemented in ARB. Bar = 10% estimated sequence divergence. GenBank accession numbers of reference sequences and clones are indicated. An asterisk indicates that the bacterium is not officially classified as an A. avenae strain. It seems likely that this strain is misnamed, because it has higher similarity with A. denitrificans type strain K601 than with the type strain of A. avenae subsp. avenae (accession number AF078759).
Benzene degradation with chlorate.
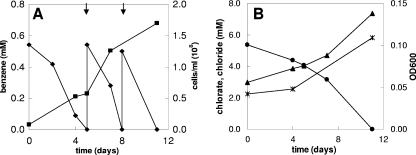
Strain BC grows on benzene with chlorate as the electron acceptor, as indicated by increases in the cell number and the OD600 (Fig. 3A and 3B). The increases in the cell number and OD600 coincided with benzene degradation. During chlorate reduction, chloride was produced. During incubation with the redox indicator resazurin, the medium turned pink due to the increased redox potential resulting from chlorate metabolism. When all of the chlorate was consumed, the medium became colorless again (after 11 days). No benzene degradation was observed in controls without inoculum or without chlorate, no chlorate reduction was observed in controls without inoculum or without benzene, and no growth was observed in controls without benzene or chlorate (results not shown). The specific growth rate of strain BC on benzene and chlorate was 0.48 day−1 (doubling time, 1.4 days). In this experiment, 0.5 mM benzene was added twice to the culture (Fig. 3A). Benzene concentrations up to 1 mM (the highest concentration tested) were degraded by strain BC. The expected stoichiometry of complete oxidation of benzene in conjunction with chlorate reduction, without taking into account the production of biomass, is:
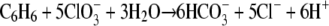 |
The experimentally measured ratio of the total amount of chlorate reduced to the amount of benzene degraded was 2.8. This ratio is lower than the expected ratio (5) according to reaction 1, but the reaction does not take into account the production of biomass. A fairly good chlorine balance was obtained; 80 to 90% of the chlorine of chlorate was recovered as chloride, indicating that no or only very small amounts of other chlorine compounds were formed. The growth yield of strain BC (based on the dry weight) determined using two cultures grown on benzene plus chlorate was 14.2 to 25.8 g of biomass per mol of benzene degraded (Table 1). Our strain produced flocculent growth on chlorate, and this may have resulted in underestimation of the growth yield due to difficulties in concentrating the cells when the dry weight method was used. To compare the biomass yield with the amount of benzene degraded and the amount of chlorate reduced, the electron equivalent mass balance approach based on the fraction of electrons from benzene used for energy production and the fraction of electrons from benzene used for cell synthesis (fs) was used as described previously (38). The cumulative amount of benzene degraded and the amount of chlorate reduced were used to calculate the fs. This calculation resulted in an fs value of about 0.39 (average of 0.34 and 0.43 [Table 1]), corresponding to the following overall balanced equation:
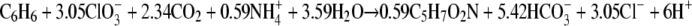 |
According to this approach, the biomass (C5H7O2N) yield should be 57.6 to 73.5 g biomass per mol benzene, which is about three times higher than the actual measured yield. In active cultures of strain BC with chlorate as the acceptor, the benzene degradation rate was about 0.35 μmol per h per mg (dry weight) biomass.
FIG. 3.
Anaerobic benzene degradation by strain BC with chlorate (5 mM) as the electron acceptor. (A) Benzene concentration (⧫) and concentration of cells (▪) at different times. (B) Chlorate (•) and chloride (▴) concentrations and OD600 (*) at different times. The arrows indicate when benzene was added.
TABLE 1.
Growth yields based on biomass determination and on the approach using the fraction of electrons from benzene used for energy production and fsa
| Expt | Amt of benzene degraded (mM) | Amt of chlorate reduced (mM) | Amt of biomass produced (mg/liter) | Yield (g cells/mol benzene) | fs | Yield based on fs (theoretical) (g cells/mol benzene) |
|---|---|---|---|---|---|---|
| 1 | 2.0 | 6.6 | 52.2 | 25.8 | 0.34 | 57.6 |
| 2 | 3.3 | 9.4 | 46.7 | 14.2 | 0.43 | 73.5 |
See reference 38.
Strain BC was also able to grow on benzene with oxygen as the electron acceptor. Different initial concentrations of oxygen were tested, including 2.5, 5, and 20% oxygen in the headspace, corresponding to nominal concentrations of 2, 4, and 16 mmol oxygen per liter, respectively (the volumes of the culture and headspace were 40 and 80 ml, respectively). Benzene degradation coincided with oxygen consumption and an increase in the OD600. The benzene degradation rates in active aerobic cultures were similar to the rates with chlorate and were not influenced by the initial oxygen concentration (results not shown). The specific growth rate of strain BC on benzene and oxygen was 0.40 day−1 (doubling time, about 1.7 days).
Strain BC was isolated in the presence of FYE, but after further transfers this bacterium grew well on benzene with chlorate in media without FYE. However, difficulties with subcultivation on benzene and chlorate occurred when outgrown cultures were stored unfed for more than 2 days. Benzene degradation ceased when benzene was not added within 1 to 2 days after the benzene was depleted. However, addition of small amounts of acetate (0.5 mM) resulted in recovery of the benzene-degrading activity (results not shown). When the cultures were degrading benzene again, benzene degradation could be sustained without addition of acetate.
Electron donors and acceptors.
All carboxylic acids tested, including acetate, lactate, pyruvate, succinate, propionate, butyrate, malate, citrate, and fumarate, were used by strain BC as sole electron donors for growth (Table 2). With carboxylic acids (10 mM) as electron donors, growth started within a few days, and all nitrate (10 mM) was consumed within 1 week. Glycerol and yeast extract were also growth substrates for strain BC. Glutamate was used by strain BC as an electron donor for nitrate reduction, but the other amino acids tested (alanine and glycine) were not used. Strain BC did not use sugars as electron donors (fructose, glucose, and xylose were tested). Within 1 week strain BC grew on the following aromatic compounds with oxygen or chlorate as the electron acceptor: benzene, toluene, phenol, o-, m-, and p-cresol, and catechol. Growth on these compounds did not occur with nitrate as the electron acceptor. Strain BC did not degrade cyclohexanol either with nitrate or with oxygen. Growth of strain BC was studied in more detail with acetate (10 mM) as the electron donor and oxygen, chlorate, or nitrate as the electron acceptor. With chlorate as the electron acceptor the growth yield appeared to be lower than the growth yield with oxygen or nitrate as the electron acceptor; i.e., lower values for cells/ml and OD600 were obtained. The specific growth rates on acetate were about 0.98, 0.56, and 0.95 day−1 with oxygen, chlorate, and nitrate as the electron acceptors, respectively.
TABLE 2.
Overview of electron donor use by strain BC
| Compound(s) | Concn used in this study | Growth with electron acceptorsa
|
||
|---|---|---|---|---|
| NO3− | O2 | ClO3− | ||
| Carboxylic acidsb | 10 mM | + | +c | +c |
| Glucose | 10 mM | − | ND | ND |
| Fructose | 10 mM | − | ND | ND |
| Xylose | 10 mM | − | ND | ND |
| Alanine | 10 mM | − | ND | ND |
| Glycine | 10 mM | − | ND | ND |
| Glutamate | 10 mM | + | ND | ND |
| Ethanol | 10 mM | − | ND | ND |
| Methanol | 10 mM | − | ND | ND |
| Glycerol | 10 mM | + | ND | ND |
| Yeast extract | 1 g/liter | + | ND | ND |
| Benzene | 0.25 mM | − | + | + |
| Toluene | 0.25 mM | − | + | + |
| Ethylbenzene | 0.25 mM | − | − | ND |
| o-Xylene | 0.1 mM | − | − | ND |
| m-Xylene | 0.1 mM | − | − | ND |
| p-Xylene | 0.1 mM | − | − | ND |
| Benzoate | 1 mM | − | − | ND |
| Phenol | 1 mM | − | + | + |
| p-Hydroxybenzoate | 1 mM | − | − | ND |
| o-Cresol | 1 mM | − | + | + |
| m-Cresol | 1 mM | − | + | + |
| p-Cresol | 1 mM | − | + | + |
| Monochlorobenzene | 0.05 mM | − | − | − |
| Catechol | 1 mM | − | + | + |
| Cyclohexanol | 1 mM | − | − | ND |
| FeCl2 | 5 mM | − | ND | ND |
| Na2S | 1 mM | − | ND | ND |
| H2d | 170 kPa | − | ND | ND |
+, growth; −, no growth; ND, not determined.
The following carboxylic acids were tested: acetate, lactate, pyruvate, succinate, propionate, butyrate, malate, citrate, and fumarate.
Only acetate was tested; the other carboxylic acids were not tested.
Acetate (1 mM) was added for heterotrophic biomass production.
In addition to chlorate and oxygen, strain BC used nitrate and nitrite. Perchlorate, sulfate, sulfite, thiosulfate, fumarate, manganese(IV) oxide, iron(III) pyrophosphate, iron(III) nitrilotriacetic acid, anthraquinone-2,6-disulfonate, bromate, selenate, and arsenate were not used as electron acceptors.
Catechol degradation.
Catechol is a central intermediate in several aerobic benzene degradation pathways (15, 20, 43). Therefore, catechol degradation by strain BC (pregrown on benzene and chlorate) was investigated with different electron acceptors. Strain BC degraded catechol with oxygen (1.2 mM) or chlorate (10 mM) as the electron acceptor but not with nitrate as the electron acceptor. The initial amount of catechol (0.8 mM) was degraded within 4 days with chlorate as the electron acceptor and within 7 days with oxygen as the electron acceptor. Controls without an inoculum showed chemical conversion of catechol (0.44, 0.25, and 0.26 mM catechol was converted in 49 days in the presence of oxygen, chlorate, and nitrate, respectively; no decrease in the oxygen, chlorate, or nitrate concentration was observed). The chemical conversion of catechol was accompanied by browning of the solution. It is known that chemical catechol polymerization can occur easily, leading to browning of a solution (40).
Detection of chlorite dismutase, benzene oxygenase, and catechol oxygenase. (i) Chlorite dismutase.
The chlorate reductase and chlorite dismutase activities in cell extracts of benzene- and chlorate-grown cells of strain BC were 0.4 and 5.7 U/mg protein, respectively. Similar chlorate reductase activity (0.3 U/mg protein) and higher chlorite dismutase activity (22 U/mg protein) were obtained with cell extracts of strain BC grown on acetate and chlorate. Gene sequences encoding chlorite dismutase (cld) were not detected in DNA extracted from strain BC by using the PCR primers used in previous studies (4). With the same primer sets we were able to detect cld genes in P. chloritidismutans (results not shown). Thus, the gene(s) encoding chlorite dismutase(s) of strain BC was too divergent from these genes to detect with the primers used.
(ii) Benzene oxygenases and extradiol dioxygenases.
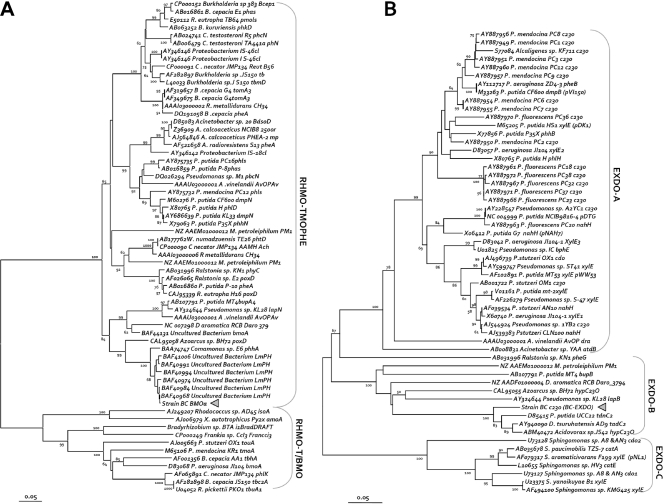
As physiological and biochemical data indicated that oxygenases are involved during growth with benzene and chlorate, we screened for the presence of fragments of genes encoding monooxygenase and dioxygenase enzymes potentially involved in benzene degradation in strain BC. No signal was detected with primers targeting genes encoding a large number of toluene/biphenyl/isopropylbenzene dioxygenases that commonly also target genes encoding benzene dioxygenases of gram-negative bacteria (58). PCR products of the expected size were detected in strain BC genomic DNA with primers sets targeting a group of type I EXDO and a group of aromatic RHMO. We confirmed the identities of the fragments by DNA sequencing, conceptual translation, and phylogenetic analyses. The translated DNA sequences encoded, in both cases, nondisrupted protein sequence frames that branched as new members of their groups (Fig. 4). In the case of the RHMO, the protein phylogeny analysis placed the large monooxygenase subunits together with the deduced amino acid sequence obtained for strain BC (BC-BMOa) (Fig. 4A) in a cluster composed of recently described members of the three-component aliphatic/aromatic monooxygenases. The closest relatives when tblastx searches (available at the NCBI website) were used were some sequences resulting from sludge amplification of community DNA from nonylphenol treatments (as referenced only in the GenBank accession number), groups of peptides of recently described variants able to monooxygenate trichloroethylene and benzene (18), and sequences detected for total DNA from enrichment experiments performed with soil samples with benzene (22). The results for the closest relative with a complete or almost complete coding DNA sequence indicated that the strain BC sequence is very similar, as assessed by using a neighbor-joining tree for a multiple-sequence alignment of proteins, to putative aromatic monooxygenase sequences found in the Comamonas sp. strain E6 (86% identity), Dechloromonas aromatica strain RCB (76% identity), and Azoarcus sp. strain BH7 (72% identity) genomes. These bacteria are all betaproteobacterial strains like strain BC and have been reported to be able to degrade monoaromatic compounds by diverse mechanisms. For instance, strain E6 is a bacterium that is able to oxygenate phenol (53).
FIG. 4.
Evolutionary relationships of the benzene monooxygenase and catechol 2,3-dioxygenase sequences found in strain BC in the contexts of the corresponding protein families. The deduced amino acid sequences for the DNA coding sequences used for primers (brackets indicate the primer pairs and clusters [see Materials and Methods]) designed to find relatives of soluble diiron monooxygenase large subunits or type I EXDO (see text), including the putative protein fragments deduced from the DNA sequences obtained from strain BC (indicated by triangles), were aligned. To generate the neighbor-joining trees shown, blocks of (on average) 220 amino acids for phenol hydroxylases and aromatic monooxygenase large subunits (A) or 237 amino acids for catechol 2,3-dioxygenases (B), spanning the length common to the gene members selected, were used (for details see Materials and Methods). Bar = 5 amino acid changes per 100 amino acids. Bootstrap values greater than 50% for 1,000 neighbor-joining trees are indicated to the left of the nodes. The primers used are indicated beside the indicated branch targeted. Sequences in the neighbor-joining trees are indicated by the DDBJ/EMBL/GenBank accession number, followed by the organism (genus or species), strain designation, and gene abbreviation.
(iii) Catechol oxygenase.
Strain BC is able to grow on catechol with oxygen and chlorate, but not with nitrate. Thus, we tried to detect genes encoding enzymes that can degrade catechol. Catechol 2,3-dioxygenases belonging to the three EXDO groups targeted (around groups I.2.A and I.2.B [16]) are expected to be present in strain BC based on the phylogenetic position of the monooxygenase sequence found in strain BC. Due to sequence variability in these clusters, primer sets able to amplify all the gene members collected, divided into three subgroups (subgroups A, B, and C), had to be designed (Fig. 4B). As predicted, amplification of strain BC genomic DNA produced a PCR product with primers targeting the group of catechol 2,3-dioxygenases from strains with monooxygenases similar to the enzyme found in strain BC. In this case, a pattern consisting of different DNA sizes, probably due to nonspecific products, was obtained due to the high degeneracy of the primers and the intentional overlapping specificity for the sequence groups targeted. A fragment of the expected product size was excised from an agarose gel and sequenced. The conceptual translation of a catechol 2,3-dioxygenase gene fragment obtained from strain BC (BC-C23O) consisted of a single reading frame without stop codons and included the conserved domains commonly found in the EXDO family, as assessed by searches of NCBI repositories. When the putative protein was aligned with the whole protein family database, the sequence was located in the middle of two experimentally proven C23O enzyme phylogenetic subgroups and in a new branch of C23O putative proteins identified in genome sequencing projects for Delftia, Azoarcus, Dechloromonas, and Pseudomonas strains (Fig. 4B).
DISCUSSION
We describe here isolation and characterization of a bacterium, strain BC, that can grow on benzene with chlorate as the electron acceptor. Strain BC is closely related to A. denitrificans type strain K601, a cyclohexanol-degrading, nitrate-reducing betaproteobacterium (99.7% similarity based on 16S rRNA genes). Besides benzene, strain BC grew on some other aromatic compounds with oxygen or in the absence of oxygen with chlorate as the electron acceptor. Strain BC is a denitrifying bacterium, but it is not able to grow on benzene with nitrate. In this respect, strain BC differs from D. aromatica strain RCB. D. aromatica strain RCB is also able to degrade ethylbenzene (9), a property that strain BC does not have. However, strain BC degrades benzene much faster and tolerates much higher benzene concentrations than D. aromatica strain RCB (8, 9, 14). Cultures of strain BC degrade about 100 μM benzene per day (Fig. 3), and this strain tolerates benzene concentrations as high as 1 mM. D. aromatica strain RCB was tested at concentrations only as high as 100 μM, and its benzene degradation rate with chlorate as the electron acceptor was less than 10 μM per day (9).
In batch cultures of strain BC growing on benzene and chlorate, the theoretical biomass yield (Table 1) was about three times higher than the actual measured yield. Low biomass production could be due to low or no energy conservation in the chlorate-to-chlorite step. So far, it is not clear how the respiratory electron transport chain is arranged and how energy is conserved in (per)chlorate-reducing bacteria (24, 60). Another possibility is that a major part of the energy is used for maintenance instead of biosynthesis. An inefficient biomass synthesis pathway or an inefficient (or incomplete) pathway for benzene degradation due to metabolite toxicity, pathway misrouting, and metabolic bottlenecks are also possible. Although no methods for CO2 production quantification were used, it is likely that strain BC degraded benzene (with oxygen or chlorate) completely to carbon dioxide. Thus, strain BC is able to grow with acetate as the electron donor, and its oxidation results inevitably in carbon dioxide. Low biomass production was also observed in other studies of anaerobic benzene degradation; e.g., the biomass production was only 2.6% for D. aromatica strain RCB when it was grown on 14C-labeled benzene and chlorate (9). Low values for incorporation of 14C-labeled benzene into biomass were also observed when strain RCB was grown with nitrate (2%) or oxygen (3%) (14).
Cell extracts of strain BC grown on benzene and on acetate with chlorate as the electron acceptor showed chlorate reductase activity (0.3 to 0.4 U/mg protein) and chlorite dismutase enzyme activity (5.7 to 22 U/mg protein), respectively. Cell extracts of P. chloritidismutans strain AW-1T had higher chlorate reductase activity (9.0 U/mg protein) and higher chlorite dismutase activity (134 U/mg protein), whereas Azospira oryzae GR-1 had chlorate reductase activity (0.39 U/mg protein) similar to that of strain BC but higher chlorite dismutase activity (145 U/mg protein) (52, 59).
During chlorate reduction, oxygen is produced by the dismutation of chlorite. Therefore, it seems likely that benzene is degraded via an aerobic degradation pathway in strain BC. Aerobic bacterial benzene degradation can be initiated by monohydroxylation or dihydroxylation. The first step in dihydroxylation is addition of dioxygen to the aromatic nucleus to form cis-benzene dihydrodiol, which is further transformed to catechol (19). Monohydroxylation is catalyzed by monooxgenases with rather broad substrate specificities; the toluene 4-monooxygenase of P. mendocina KR1, the toluene 3-monooxygenase of Ralstonia pickettii PKO1, and the toluene ortho-monooxygenase of Burkholderia cepacia G4 all convert benzene to phenol, as well as catechol and 1,2,3-trihydroxybenzene in successive hydroxylation reactions (46). Strain BC degrades phenol and catechol with oxygen and chlorate as the electron acceptors but not with nitrate. In addition, chlorate reductase and chlorite dismutase activities were found in cell extracts of strain BC. An aerobic degradation pathway normally requires chlorate reductase, chlorite dismutase, and oxygenase enzymes.
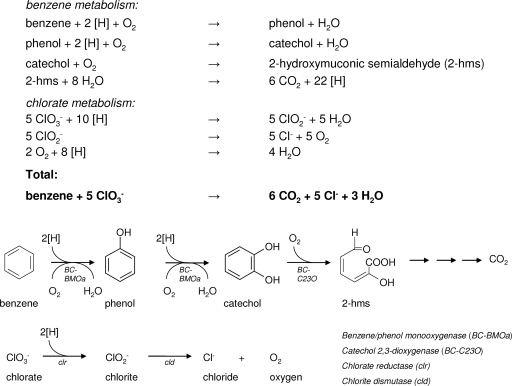
While it is obvious that additional experiments are needed to precisely define the activities of the oxygenase systems encoded in the genome of strain BC, based on the physiological, genetic, and biochemical experiments presented in this study, we propose a benzene degradation pathway with chlorate as the electron acceptor in strain BC (Fig. 5). In this pathway, oxygen produced during chlorate reduction is used in oxygenase reactions; i.e., benzene is converted to catechol by two sequential monooxygenase (benzene monooxygenase [BC-BMOa]) reactions, and catechol is converted to 2-hydroxymuconic semialdehyde by catechol 2,3-dioxygenase (BC-C23O). The possibility of dihydroxylation of benzene to catechol cannot be ruled out, but genes encoding benzene dioxygenases were not detected with the primers used in this study. Electrons (reducing equivalents) for chlorate reduction must be derived from intermediates of the aerobic benzene degradation pathway. This pathway could probably explain the difficulties experienced with subcultivation on benzene and chlorate when cultures of strain BC were stored unfed. Oxygen and reducing equivalents are required for benzene degradation by means of oxygenases, while reducing equivalents are also needed to initiate chlorate reduction. Apparently, in starved cells the lack of reducing equivalents causes problems for initiation of the metabolism and growth of strain BC. Addition of an easily degradable substrate (e.g., FYE or acetate) results in the onset of growth and benzene degradation. We found that addition of oxygen was not sufficient to initiate benzene degradation, which is another indication of the involvement of (mono)oxygenases.
FIG. 5.
Proposed benzene degradation pathway with chlorate as the electron acceptor in strain BC and proposed stoichiometric reactions involved in benzene degradation with chlorate as the electron acceptor in strain BC. [H] indicates reducing equivalents. Benzene metabolism involves the hydroxylation of benzene to phenol, phenol hydroxylation to catechol, extradiol (meta) cleavage of catechol to 2-hydroxymuconic semialdehyde (2-hms), and complete oxidation of 2-hydroxymuconic semialdehyde to carbon dioxide and reducing equivalents. Chlorate metabolism involves the reduction of chlorate to chlorite, dismutation of chlorite into chloride and oxygen, and subsequent reduction of oxygen to water.
In conclusion, the results of the physiological, genetic, and biochemical experiments performed in this study strongly suggest that oxygen formed during the dismutation of chlorite may be used not only as the terminal electron acceptor in strain BC but also to attack molecules by means of oxygenases. This demonstrates that there are aerobic benzene bacterial biodegradation pathways under essentially anaerobic conditions involving the concerted action of chlorite dismutases, providing the metabolic oxygen needed by aromatic activating and cleaving oxygenases. Thus, aerobic aromatic degradation pathways can be employed by single organisms under conditions in which no external oxygen is supplied.
Acknowledgments
We thank Farai Maphosa for his input and help with construction of the phylogenetic tree based on partial 16S rRNA gene sequences. We thank Dietmar H. Pieper for his valuable insights regarding putative oxygenase gene screening for isolate BC and Silke Kahl for her excellent technical assistance.
This research was financed through grants from the WIMEK graduate school (Wageningen Institute for Environment and Climate Research) (www.dow.wau.nl/msa/wimek and www.sense.nl) and from SKB (Dutch Center for Soil Quality Management and Knowledge Transfer) (www.skbodem.nl), and it was incorporated into the TRIpartite Approaches toward Soil Systems Processes (TRIAS) program (www.nwo.nl/trias). H. Junca thanks the European Commission for providing financial support through contract 003998 (GOCE) to the Biotool project (www.gbf.de/biotools).
Footnotes
Published ahead of print on 12 September 2008.
REFERENCES
- 1.Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baek, S. H., K. H. Kim, C. R. Yin, C. O. Jeon, W. T. Im, K. K. Kim, and S. T. Lee. 2003. Isolation and characterization of bacteria capable of degrading phenol and reducing nitrate under low-oxygen conditions. Curr. Microbiol. 47:462-466. [DOI] [PubMed] [Google Scholar]
- 4.Bender, K. S., M. R. Rice, W. H. Fugate, J. D. Coates, and L. A. Achenbach. 2004. Metabolic primers for detection of (per)chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Appl. Environ. Microbiol. 70:5651-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blümel, S., H. J. Busse, A. Stolz, and P. Kämpfer. 2001. Xenophilus azovorans gen. nov., sp. nov., a soil bacterium that is able to degrade azo dyes of the Orange II type. Int. J. Syst. Evol. Microbiol. 51:1831-1837. [DOI] [PubMed] [Google Scholar]
- 6.Botton, S., and J. R. Parsons. 2007. Degradation of BTX by dissimilatory iron-reducing cultures. Biodegradation 18:373-381. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty, R., and J. D. Coates. 2005. Hydroxylation and carboxylation—two crucial steps of anaerobic benzene degradation by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:5427-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty, R., S. M. O'Connor, E. Chan, and J. D. Coates. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:8649-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, W., Y. Um, and T. R. P. Holoman. 2005. Molecular characterization of anaerobic microbial communities from benzene-degrading sediments under methanogenic conditions. Biotechnol. Prog. 21:1789-1794. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. C., J. K. Hwang, and J. M. Yang. 2006. (PS)2: protein structure prediction server. Nucleic Acids Res. 34:W152-W157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coates, J. D., and L. A. Achenbach. 2004. Microbial perchlorate reduction: rocket-fuelled metabolism. Nat. Rev. Microbiol. 2:569-580. [DOI] [PubMed] [Google Scholar]
- 13.Coates, J. D., R. A. Bruce, J. Patrik, and L. A. Achenbach. 1999. Hydrocarbon bioremediative potential of perchlorate-reducing bacteria. Bioremediat. J. 3:323-334. [Google Scholar]
- 14.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039-1043. [DOI] [PubMed] [Google Scholar]
- 15.Dagley, S. 1985. Microbial metabolism of aromatic compounds, p. 483-505. In M. Moo-Young (ed.), The principles of biotechnology, vol. I. Pergamon Press, Oxford, United Kingdom. [Google Scholar]
- 16.Eltis, L. D., and J. T. Bolin. 1996. Evolutionary relationships among extradiol dioxygenases. J. Bacteriol. 178:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, A., I. Herklotz, S. Herrmann, M. Thullner, S. A. B. Weelink, A. J. M. Stams, H. H. Richnow, and C. Vogt. 2008. Combined carbon and hydrogen isotope fractionation investigations for elucidating benzene biodegradation pathways. Environ. Sci. Technol. 42:4356-4363. [DOI] [PubMed] [Google Scholar]
- 18.Futamata, H., S. Harayama, and K. Watanabe. 2001. Group-specific monitoring of phenol hydroxylase genes for a functional assessment of phenol-stimulated trichloroethylene bioremediation. Appl. Environ. Microbiol. 67:4671-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, D. T., J. R. Koch, and R. E. Kallio. 1968. Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry 7:2653-2662. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, D. T., and V. Subramanian. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-252. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, Inc., New York, NY.
- 21.Holliger, C., G. Schraa, A. J. M. Stams, and A. J. B. Zehnder. 1993. A highly purified enrichment culture couples the reductive dechlorination of tetrachloroethene to growth. Appl. Environ. Microbiol. 59:2991-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwai, S., F. Kurisu, H. Urakawa, O. Yagi, and H. Furumai. 2007. Development of a 60-mer oligonucleotide microarray on the basis of benzene monooxygenase gene diversity. Appl. Microbiol. Biotechnol. 75:929-939. [DOI] [PubMed] [Google Scholar]
- 23.Kasai, Y., Y. Takahata, M. Manefield, and K. Watanabe. 2006. RNA-based stable isotope probing and isolation of anaerobic benzene-degrading bacteria from gasoline-contaminated groundwater. Appl. Environ. Microbiol. 72:3586-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kengen, S. W. M., G. B. Rikken, W. R. Hagen, C. G. van Ginkel, and A. J. M. Stams. 1999. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J. Bacteriol. 181:6706-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 26.Kunapuli, U., C. Griebler, H. R. Beller, and R. U. Meckenstock. 2008. Identification of intermediates formed during anaerobic benzene degradation by an iron-reducing enrichment culture. Environ. Microbiol. 10:1703-1712. [DOI] [PubMed] [Google Scholar]
- 27.Leahy, J. G., P. J. Batchelor, and S. M. Morcomb. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27:449-479. [DOI] [PubMed] [Google Scholar]
- 28.Logan, B. E. 1998. A review of chlorate- and perchlorate-respiring microorganisms. Bioremed. J. 2:69-79. [Google Scholar]
- 29.Logan, B. E., and J. Wu. 2002. Enhanced toluene degradation under chlorate-reducing conditions by bioaugmentation of sand columns with chlorate- and toluene-degrading enrichments. Bioremed. J. 6:87-95. [Google Scholar]
- 30.Lovley, D. R. 2000. Anaerobic benzene degradation. Biodegradation 11:107-116. [DOI] [PubMed] [Google Scholar]
- 31.McGinnis, S., and T. L. Madden. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20-W25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mechichi, T., E. Stackebrandt, and G. Fuchs. 2003. Alicycliphilus denitrificans gen. nov., sp. nov., a cyclohexanol-degrading, nitrate-reducing beta-proteobacterium. Int. J. Syst. Evol. Microbiol. 53:147-152. [DOI] [PubMed] [Google Scholar]
- 33.Musat, F., and F. Widdel. 2008. Anaerobic degradation of benzene by a marine sulfate-reducing enrichment culture, and cell hybridization of the dominant phylotype. Environ. Microbiol. 10:10-19. [DOI] [PubMed] [Google Scholar]
- 34.Nicholas, K. B., H. B. J. Nicholas, and D. W. I. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 35.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 36.Plugge, C. M., E. G. Zoetendal, and A. J. M. Stams. 2000. Caloramator coolhaasii sp. nov., a glutamate-degrading, moderately thermophilic anaerobe. Int. J. Syst. Evol. Microbiol. 50:1155-1162. [DOI] [PubMed] [Google Scholar]
- 37.Rikken, G. B., A. G. M. Kroon, and C. G. van Ginkel. 1996. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 45:420-426. [Google Scholar]
- 38.Rittmann, B. E., and P. L. McCarty. 2001. Environmental biotechnology: principles and applications. McGraw-Hill Book Company, New York, NY.
- 39.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Cortes, S., O. Francioso, J. V. Garcia-Ramos, C. Ciavatta, and C. Gessa. 2001. Catechol polymerization in the presence of silver surface. Colloids Surf. A 176:177-184. [Google Scholar]
- 41.Sanguinetti, C. J., E. D. Neto, and A. J. G. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:914-921. [PubMed] [Google Scholar]
- 42.Scholten, J. C. M., and A. J. M. Stams. 1995. The effect of sulfate and nitrate on methane formation in a freshwater sediment. Antonie van Leeuwenhoek 68:309-315. [DOI] [PubMed] [Google Scholar]
- 43.Smith, M. R. 1990. The biodegradation of aromatic hydrocarbons by bacteria. Biodegradation 1:191-206. [DOI] [PubMed] [Google Scholar]
- 44.Stams, A. J. M., J. B. Vandijk, C. Dijkema, and C. M. Plugge. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl. Environ. Microbiol. 59:1114-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan, N. C. G., W. van Doesburg, A. A. M. Langenhoff, and A. J. M. Stams. 2006. Benzene degradation coupled with chlorate reduction in a soil column study. Biodegradation 17:113-119. [DOI] [PubMed] [Google Scholar]
- 46.Tao, Y., A. Fishman, W. E. Bentley, and T. K. Wood. 2004. Oxidation of benzene to phenol, catechol, and 1,2,3-trihydroxybenzene by toluene 4-monooxygenase of Pseudomonas mendocina KR1 and toluene 3-monooxygenase of Ralstonia pecketii PKO1. Appl. Environ. Microbiol. 70:3814-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulrich, A. C., H. R. Beller, and E. A. Edwards. 2005. Metabolites detected during biodegradation of 13C6-benzene in nitrate-reducing and methanogenic enrichment cultures. Environ. Sci. Technol. 39:6681-6691. [DOI] [PubMed] [Google Scholar]
- 49.Ulrich, A. C., and E. A. Edwards. 2003. Physiological and molecular characterization of anaerobic benzene-degrading mixed cultures. Environ. Microbiol. 5:92-102. [DOI] [PubMed] [Google Scholar]
- 50.Urbansky, E. T. 1998. Perchlorate chemistry: implications for analysis and remediation. Bioremediat. J. 2:81-95. [Google Scholar]
- 51.van Doesburg, W., M. H. A. van Eekert, P. J. M. Middeldorp, M. Balk, G. Schraa, and A. J. M. Stams. 2005. Reductive dechlorination of beta-hexachlorocyclohexane (beta-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 54:87-95. [DOI] [PubMed] [Google Scholar]
- 52.Van Ginkel, C. G., G. B. Rikken, A. G. M. Kroon, and S. W. M. Kengen. 1996. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 166:321-326. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe, K., S. Hino, K. Onodera, S. Kajie, and N. Takahashi. 1996. Diversity in kinetics of bacterial phenol-oxygenating activity. J. Ferment. Bioeng. 81:560-563. [Google Scholar]
- 54.Weelink, S. A. B., N. C. G. Tan, H. ten Broeke, W. van Doesburg, A. A. M. Langenhoff, J. Gerritse, and A. J. M. Stams. 2007. Physiological and phylogenetic characterization of a stable benzene-degrading chlorate-reducing microbial community. FEMS Microbiol. Ecol. 60:312-321. [DOI] [PubMed] [Google Scholar]
- 55.Wen, A. M., M. Fegan, C. Hayward, S. Chakraborty, and L. I. Sly. 1999. Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:567-576. [DOI] [PubMed] [Google Scholar]
- 56.Willems, A., J. Deley, M. Gillis, and K. Kersters. 1991. Comamonadaceae, a new family encompassing the acidovorans ribosomal RNA complex, including Variovorax paradoxus gen. nov., comb. nov., for Alcaligenes paradoxus (Davis 1969). Int. J. Syst. Bacteriol. 41:445-450. [Google Scholar]
- 57.Willems, A., M. Goor, S. Thielemans, M. Gillis, K. Kersters, and J. Deley. 1992. Transfer of several phytopathogenic Pseudomonas species to Acidovorax as Acidovorax avenae subsp. avenae subsp. nov., comb. nov., Acidovorax avenae subsp. citrulli, Acidovorax avenae subsp. cattleyae, and Acidovorax konjaci. Int. J. Syst. Bacteriol. 42:107-119. [DOI] [PubMed] [Google Scholar]
- 58.Witzig, R., H. Junca, H. J. Hecht, and D. H. Pieper. 2006. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: links between benzene biodegradation and genes similar to those encoding isopropylbenzene dioxygenases. Appl. Environ. Microbiol. 72:3504-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolterink, A. F. W. M., A. B. Jonker, S. W. M. Kengen, and A. J. M. Stams. 2002. Pseudomonas chloritidismutans sp. nov., a nondenitrifying, chlorate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 52:2183-2190. [DOI] [PubMed] [Google Scholar]
- 60.Wolterink, A. F. W. M., E. Schiltz, P. L. Hagedoorn, W. R. Hagen, S. W. M. Kengen, and A. J. M. Stams. 2003. Characterization of the chlorate reductase from Pseudomonas chloritidismutans. J. Bacteriol. 185:3210-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]