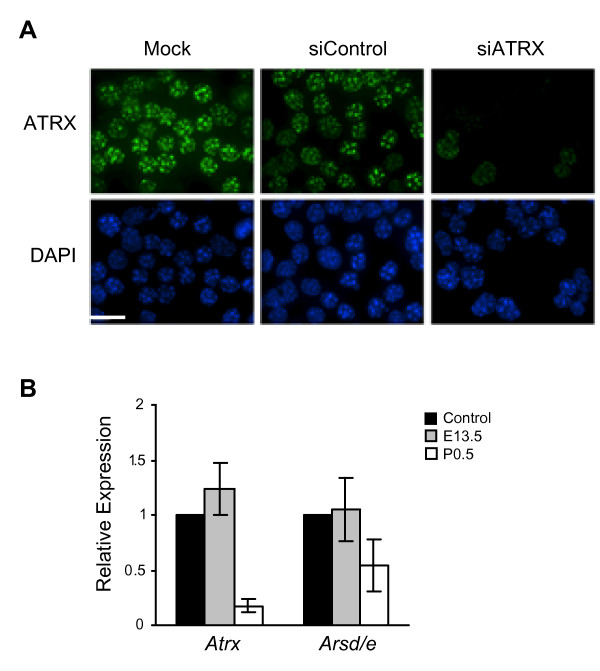
Figure 4.
Arsd/e transcriptional downregulation is recapitulated in ATRX-depleted cells. (A) RNA interference was used to deplete ATRX in Neuro-2a neuroblastoma cells. Cells were transfected with 8 nM siRNA, fixed after 72 h and processed for immunofluorescence staining using an anti-ATRX primary antibody (H300) and anti-rabbit Alexa 488 secondary antibody, then counterstained with DAPI to detect nuclei. In the siATRX treated samples, approximately 95% of cells were negative for ATRX. Scale bar = 20 μM. (B) Total RNA was isolated for quantitative real-time PCR of Atrx and Arsd/e gene expression at 72 hours post-transfection. Mock (transfection reagent only) expression levels were set to one and a non-specific siRNA was used as a control. Results were normalized to β-actin expression levels. Error bars represent standard error of the mean (n = 3).