Fig. 5.
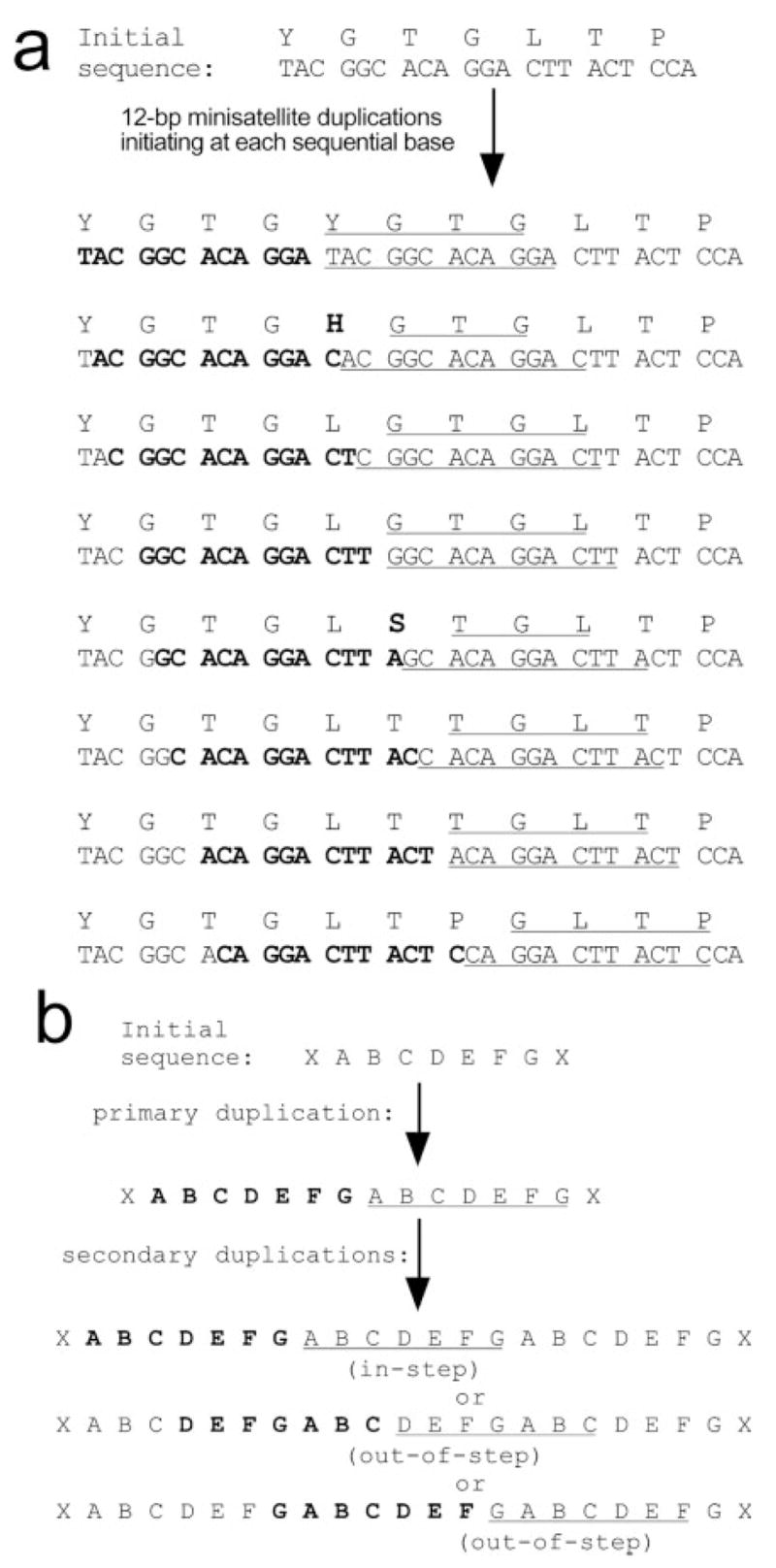
Models of minisatellite-dependent, oligopeptide-encoding sequence duplications. (a) Reading-frame independent preservation of amino acid sequences. A 7-aa region of the mouse NN region (top) was chosen as a starting sequence; however similar results are obtained with most arbitrary sequences (not shown). Twelve–base pair minisatellite duplications were modeled starting at each of eight consecutive nucleotides. Duplicated nucleotides are designated by bold font and underlining. Duplicated amino acids are underlined; novel amino acids are in bold. Few duplications (two of eight depicted) yield a novel amino acid, and this only occurs at the template/copy junction. (b) Within repeats, duplications preserve repeat periodicity. Model shows duplication of a 7-aa motif, “ABCDEFG” (21-bp minisatellite). Bold and underline indicate duplicated regions. Secondary duplications that initiate either at the first amino acid within the repeat (residue A, “in-step”) or at any other amino acid within the repeat (residues D and G shown, “out-of-step”) lead to the same trimeric repeat: ABCDEFGABCDEFGABCDEFG. The models in (a) and (b) show that imprecise or arbitrary duplications, as long as they occur in multiples of 3 bp, will tend to reiterate existing amino acid sequences and repeat motifs, rather than to insert novel amino acids or disrupt repeat units.
