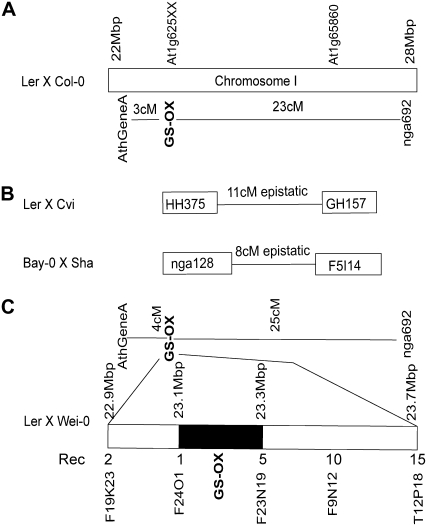
Figure 3.
Genetic mapping of GS-OX in multiple Arabidopsis RIL populations. The position of the GS-OX loci [controlling MT:(MT + MS) ratio] on chromosome I in four different Arabidopsis RIL populations is presented. The genomic position of At1g65860 (FMOGS-OX1) and At1g62540 to At1g62580 (At1g625XX) is presented for reference. A, For the Ler × Col-0 population, GS-OX was mapped as a qualitative locus and the position is presented in relation to the AthGeneA and nga692 markers. B, In Ler × Cvi and Bay-0 × Sha, GS-OX was mapped as a quantitative trait and the one LOD intervals for the identified loci on chromosome I presented with the genetic distance between them. The tested marker for each QTL is positioned relative to its physical position on chromosome I. Epistatic shows that there was a significant interaction between these two QTLs. C, For the Ler × Wei-0 F2 population, GS-OX was mapped as a qualitative locus and the position is presented in relation to the AthGeneA and nga692 markers. The Ler × Wei-0 population was used to fine map the major GS-OX locus and the fine-mapping information is presented at the bottom with the genomic position of the markers indicated. Rec, Number of recombinations between the given marker and the GS-OX phenotype in 400 F2 individuals. No markers were identified that could further resolve the position of the GS-OX locus as indicated by the black box.