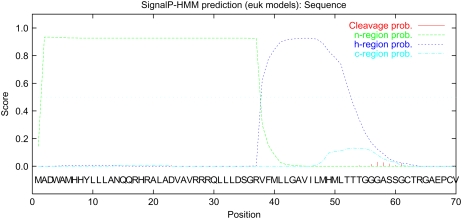
Figure 1.
Output of SignalP-HMM prediction of XA27 from the SignalP 3.0 server. A signal-anchor-like sequence was predicted at the N-terminal region of XA27, which contains an N-terminal positively charged region (n-region; residues 1–37) followed by a hydrophobic region (h-region; residues 38–57; Emanuelsson et al., 2007). The full-length amino acid sequence of XA27 was submitted to the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) for prediction of the signal peptide or signal anchor of eukaryotes. Methods of both neural networks and hidden Markov models were used for prediction. Standard output format was selected.