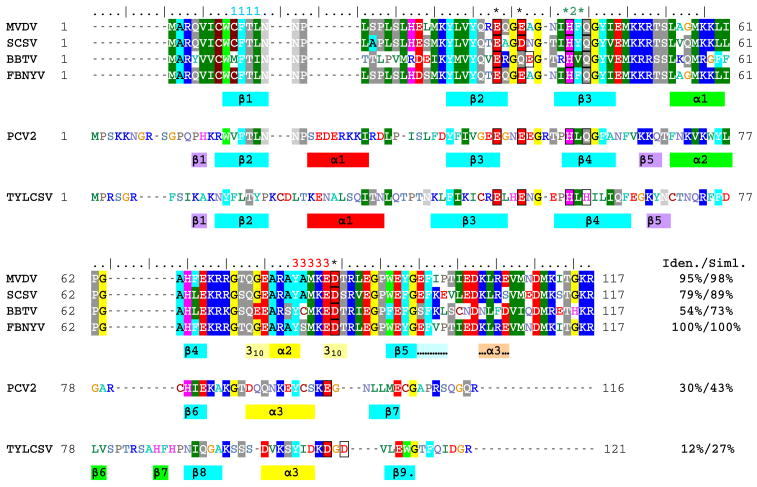
FIGURE 7.
Structure-based alignment of the Rep endonuclease domain sequences from the nanoviruses faba bean necrotic yellows virus (FBNYV, accession number O39828), milk vetch dwarf virus (MVDV, NP_619769), subterranean clover stunt virus (SCSV, NP_620694), and banana bunchy top virus (BBTV, NP_604483)), the circovirus porcine circovirus type 2 (PCV2, AAQ94098)), and the geminivirus tomato yellow leaf curl Sardinia virus (TYLCSV, CAA43466). The secondary structure elements present in FBNYV M-Rep2-95 (identical color coding as in Figure 3B), as well as those in the extension Tag21-M-Rep1-.117 (indicated with “…” and highlighted in pale blue and orange), together with those of the previously determined structures of the Rep endonuclease domains of PCV2 (20) and TYLCSV (13) are displayed below the corresponding sequences. The conserved RCR initiator protein sequence motifs 1, 2, and 3 are labeled above the alignment by 1111, *2*, and 33333, respectively. Putative divalent metal binding residues are boxed and labeled above the alignment by an asterisk. The pale yellow bars flanking helix α2 in FBNYV indicate regions adopting a 310 helix conformation (see text). Sequence identities (Iden.) and similarities (Siml.) of the different sequences compared to the FBNYV Rep sequence are provided at the end of the alignment.