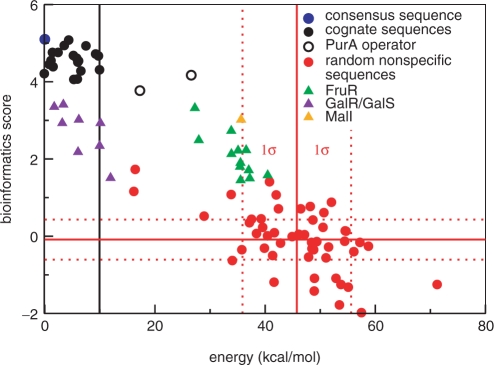
Figure 2.
Bioinformatics score versus energy. All binding energies are shown relative to the binding energy of the consensus sequence seqc (blue circle) at 0 kcal/mol. Black circles: 21 binding sequences selected by Mironov et al. (14), PurA sequences are unfilled. Red circles: random noncognate sequences selected from the E. coli genome. Green, indigo and orange triangles: FruR, GalR/GalS and MalI operator sites. The solid red lines indicate the average energy or average bioinformatics score for the random sequences; the dashed lines mark the first SD. The solid black line goes through the data point for the third worst cognate sequence (a black circle). The two cognate sequences with even worse binding energies (hollow black circle) are controversial binding sites. The linear correlation coefficient is −0.6 for the random sequences and −0.8 for all sequences displayed.