Abstract
Actin is a major cytoskeletal protein in eukaryotes. Recent studies suggest more diverse functional roles for this protein. Actin mRNA is known to be localized to neuronal synapses and undergoes rapid deadenylation during early developmental stages. However, its 3′-untranslated region (UTR) is not characterized and there are no experimentally determined polyadenylation (polyA) sites in actin mRNA. We have found that the cytoplasmic β-actin (Actb) gene generates two alternative transcripts terminated at tandem polyA sites. We used 3′-RACE, EST end analysis and in situ hybridization to unambiguously establish the existence of two 3′-UTRs of varying length in Actb transcript in mouse neuronal cells. Further analyses showed that these two tandem polyA sites are used in a tissue-specific manner. Although the longer 3′-UTR was expressed at a relatively lower level, it conferred higher translational efficiency to the transcript. The longer transcript harbours a conserved mmu-miR-34a/34b-5p target site. Sequence-specific anti-miRNA molecule, mutations of the miRNA target region in the 3′-UTR resulted in reduced expression. The expression was restored by a mutant miRNA complementary to the mutated target region implying that miR-34 binding to Actb 3′-UTR up-regulates target gene expression. Heterogeneity of the Actb 3′-UTR could shed light on the mechanism of miRNA-mediated regulation of messages in neuronal cells.
INTRODUCTION
Actin is an abundantly expressed globular structural protein found in eukaryotes, involved in muscle contraction, cell mobility, cytokinesis, organelle movement and maintenance of cell shape. In lower organisms like yeast, actin is coded for by a single gene whereas different isoforms of actin perform different functions in higher organisms like mammals. The expression of cytoplasmic β-actin gene is generally believed to be constitutive and ubiquitous and it is well known that it carries out housekeeping functions in the eukaryotic cell. However, recently, actin has been shown to play diverse roles besides its housekeeping functions. Presence of actin in the nucleus has been demonstrated and functionally, it has been shown to interact with the transcriptional machinery, suggesting a role in transcriptional regulation (1–5). In several cell types, including chicken fibroblasts and mammalian neuronal cells, actin mRNA is known to be selectively localized to the periphery of the cell. The high conservation of the 3′-untranslated region (UTR) of actin in vertebrates led to the suggestion that they may contain regulatory sequences (6). Subsequently, it has been demonstrated that a 54-nucleotide element called the zipcode, immediately following the stop codon is necessary for the localization of actin mRNA (7). Actin is also known to be deadenylated during early developmental stages along with many other transcripts (8,9). Besides its role in localization, the 3′-UTR of the actin mRNA has not been extensively studied and barring EST-based evidence, there exists no experimentally determined polyadenylation (polyA) site in the actin transcript.
The polyA of eukaryotic transcripts is a two-step enzymatically driven process, wherein mRNA is cleaved at a specific site and subsequently, the action of polyA polymerase, in the presence of a large basal polyA machinery, adds adenosine nucleotides to the mRNA (10–16). Several factors like the cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF) and cleavage factors (CFs I and II) perform specific functions within the polyA machinery. The polyA is important in conferring transcript stability (17,18) and translational efficiency (19–23). The polyA tail is also believed to target the RNA for nuclear export (24,25). Although the enzymatic process of polyA is well understood, the sequences in the mRNA that specify the site of cleavage and extent of polyA are not fully characterized. In a majority of cases, cis-acting elements surrounding the polyA site act together to specify the cleavage site. The sequence motif AAUAAA referred to as polyadenylation signal (PAS) that occurs about 10–30 nt (26,27) upstream of the polyA site in majority of human transcripts (28,29), binds to the 160 kDa subunit of CPSF. However, variants of the AAUAAA motif can effectively drive polyA (28,29). Besides the PAS element, G/U or U rich sequences downstream to the PAS contribute to the polyA site (30–32) by binding to the multi-protein CstF.
Recently, bioinformatic analyses of the occurrence of polyA sites in the human and mouse genomes have shown a surprisingly large number of alternative polyA sites (28,33). Large-scale mapping of EST sequences to the human and mouse genome showed that alternative polyadenylation is widespread in the mouse and human genomes. The hexamer motifs present upstream of the polyA sites identified by EST analysis vary significantly from the canonical motif, AAUAAA. Ten to thirteen variants of the PAS element account for the polyA sites identified over the whole genome. The PAS hexamers AAUAAA and AUUAAA together account for nearly 69% of all polyA sites in the human genome (28). The length of the 3′-UTR, defined by the distance between the stop codon and the proximal polyA site was highly variable with a median length of 324 in humans and 385 nt in mouse (28). The frequent occurrence of polyA sites within internal exons and introns of genes implies that the use of alternative polyA sites contributes significantly to the complexity of the eukaryotic transcriptome. When the alternative sites occur within protein-coding regions of the transcript, it can result in functionally different products belonging to the same gene family, as shown in the case of the Lamin genes (33). Transcripts coding for the same protein but containing different 3′-UTRs may differ in stability, localization, translational efficiency and targeting by small regulatory RNAs. Such cases called tandem polyA sites (34) have been shown in several genes and implicated in differential expression of transcripts in tissues. For instance, the transcripts from the huntingtin gene, that causes the Huntington's neurodegenerative disorder, are preferentially polyadenylated at the distal site in the brain, but truncated at the proximal polyA site in other tissues (35). Testis, is one of the tissues in which frequent use of alternative tandem polyA site has been observed (34). At least two groups have independently reported the biased use of alternative polyA sites in different tissues. By mapping ESTs from libraries derived from different tissues, Beaudoing et al. (36) showed that polyA usage is biased in a tissue- and disease-related manner. Zhang et al. (37) have also shown that the usage of polyA sites as well as expression of protein factors involved in polyA is likely to be different in brain tissues compared to other tissues (37). In spite of the evidence from large-scale analysis of EST sequences, characterization of polyA sites and 3′-UTR variation in mammalian transcripts requires detailed experimental analysis. Experimental proof is lacking for majority of the predicted alternative polyA events.
Here we used clues from hybridization patterns of probe sets in high-density oligonucleotide (Affymetrix, CA, USA) arrays to identify potentially differentially expressed transcripts from the actin gene and confirmed it with experimental analysis and EST mapping. Further analysis of microarray data and a survey of EST sequences showed that the cytoplasmic β-actin gene in mouse may use two tandem polyA sites that are used in a tissue-specific manner. We carried out 3′-RACE analysis to unambiguously establish the existence of two 3′-UTRs of varying length in actin transcripts. The distal polyA site is associated with a perfect PAS element (AAUAAA). Although a U-rich potential CstF-binding site is present downstream to the proximal PAS, no upstream canonical PAS element could be identified. On the other hand although the longer UTR-containing transcript was expressed at a relatively lower level in neuronal cells, it conferred higher translational efficiency to the transcript and harbours miRNA target sites. Post-transcriptional regulation of expression from the longer UTR-containing transcript is mediated, at least in part by a conserved mmu-miR-34b-5p/34a target site. LNA-modified anti-miRNA against the miRNA, mutations in the miRNA seed pairing region in the UTR resulted in down-regulation of the target suggesting that the miRNA enhances the target expression. The expression of the target was restored using a mutant miRNA complementary to the mutant target region. Here we provide the first evidence for differential regulation of alternative transcripts through miRNA-mediated up-regulation.
MATERIALS AND METHODS
EST and microarray data analysis
Microarray data [accession id: E-HGMP-2, Freilich et al. (38)] were downloaded from Array express site. Data were normalized by using Z-score transformation method as described (39).
Mouse ESTs were downloaded from the Unigene (IDs Mm.455830, Mm.391967, Mm.328431) and comprised of a total of 5195 ESTs derived from 43 tissues. 3′- Read ESTs with PAS were parsed using Perl scripts as per the Unigene annotations. The final dataset of 3′-read ESTs with PASs comprised of 203 unique entries. The EST sequences were mapped back to the genome location using BLAT using default options. The EST end positions and the tissues from which they were derived was parsed using Perl scripts with reference to the gene structure and positions (UCSC mm9).
RNA isolation
Total RNA was isolated from cultured Neuro-2a cells using TRIzol reagent (Invitrogen, USA) according to the manufacturer's instruction. The RNA pellets were washed with 70% ethanol, centrifuged and dried. Pellets were re-suspended in Diethyl pyrocarbonate-treated water followed by the addition of 10× reaction buffer and RNAse free DNase I (Fermentas, Germany). Samples were incubated at 37°C for 30 min. Then the RNA was cleaned using RNeasy Mini Kit (Qiagen, Hilden, Germany) following the protocol by the manufacturer. RNA concentration and purity were determined by measuring optical density at 260 and 280 nm using a spectrophotometer (Eppendorf, Hamburg, Germany) and separating the RNA samples on a 1.5% agarose formaldehyde gel.
Rapid amplification of 3′-cDNA ends (3′-RACE)
cDNA was generated from total RNA sample by using adapter primer [AP: GGCCACGCGTCGACTAGTAC (T)17] and M-MuLV reverse transcriptase (New England Biolabs Inc., UK) at 42°C for 1 h. Subsequently RNaseH (Invitrogen) treatment was performed at 37°C for 20 min.
Touch down (TD) PCR was carried out using the following primers:
GGCATTGCTGACAGGATGCAGAAGG (gene-specific primer)
GGCCACGCGTCGACTAGTAC (same as AP primer except T17 unit)
TD PCR condition was as followed: 94°C for 3 min; 29 cycles of 94°C for 1 min, 65°C (with reduction per cycle 0.5°C) for 2 min, 72°C (with elevation per cycle 0.2°C) for 1 min; then 9 cycles of 94°C for 1 min, 55°C for 2 min and 72°C for 2 min; then final extension of 72°C for 7 min.
Nested PCR was performed by using primer 2 (above) and the primer below:
GAAGGAGATTACTGCTCTGGCTCC [Nested primer for primer (1)]. Nested PCR condition: 95°C for 5 min, 25 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 2 min, then final extension at 72°C for 10 min. In this way, we were able to detect short 3′-UTR.
Long 3′-UTR was amplified by using TD PCR product as template and Primer 2 (above) and long UTR-specific primer mentioned below:
GTTTTGGCGCTTTTGACTCAGGATT
PCR condition was 95°C for 5 min, 35 cycles of 95°C for 10 s, 60°C for 30 s, 72°C for 30 s, then final extension at 72°C for 5 min.
PCR products were resolved on a EtBr-stained 2% agarose gel. Purified PCR products were cloned in T-vector (pCR4®-TOPO®, Invitrogen) and confirmed by sequencing.
Constructs
Cloning of long- and short-UTR of Actb
The short and long 3′-UTR of murine Actb were amplified from 3′-RACE product synthesized from Neuro-2a cells. The following primers introduced HindIII (primer 1) and SacI sites (Primer 2 and 3) in the PCR products (restriction site underlined):
Reverse primer: AAAAAGCTTGGCCACGCGTCGACTAGTAC (common for long and short, contains sequence of AP)
Forward primer: AAAGAGCTCGTTTTGGCGCTTTTGACTCAGGATT (for long)
Forward primer: AAAGAGCTCATCGTGCACCGCAAGTGCTTC (for short)
The gel purified PCR products were cloned in HindIII and SacI site of pMIR-REPORTTM vector (Ambion, Texas, USA), in frame with luciferase and confirmed by sequencing.
Cloning of long UTR in antisense orientation
The long UTR sequence was amplified from long UTR construct (mentioned above) using the following primers:
Forward primer: AAAAAGCTTGTTTTGGCGCTTTTGACTCAGGATT (HindIII recognition sequence underlined).
Reverse primer: AAAGAGCTCGGCCACGCGTCGACTAGTAC (SacI recognition sequence underlined).
The gel purified PCR product was cloned in HindIII and SacI site of pMIR-REPORTTM vector (Ambion), in frame with luciferase and confirmed by sequencing.
Cloning of mutant versions of long UTR
Mutant versions (named mut1 and mut2) of long 3′-UTR of Actb (long UTR) were prepared by introducing two base changes to the miRNA (mmu-miR-34a/b-5p) seed paring region. The following primers were used for preparing mut1:
FP1: AAAGAGCTCGTTTTGGCGCTTTTGACTCAGGATT (SacI site underlined).
RP1: AAGTCACTCTACAGGCCAGCCCTGGCTGCC (mutant bases underlined).
FP2: GGCCTGTAGAGTGACTTGAGACCAATAAAAGTGCA (mutant bases underlined).
RP2: AAAAAGCTTGGCCACGCGTCGACTAGTAC (HindIII site underlined). [Note that primer (2) and (3) have overlapping regions that include mutant bases.]
PCR reactions were performed using long UTR construct (mentioned above) as a template. We obtained PCR product I (482 bp) using primer (1) and (2) and PCR product II (93 bp) by using primer (3) and (4). PCR products I and II were then heated at 95°C for 20 min and annealed at room temperature for 20 min. Annealed product was then used as a template and amplified by using primer (1) and (4). The gel purified PCR product was cloned in HindIII and SacI site of pMIR-REPORT™ vector (Ambion), in frame with luciferase and confirmed by sequencing.
For creating mut2, the following primers were used:
FP1: AAAGAGCTCGTTTTGGCGCTTTTGACTCAGGATT (SacI site underlined)
RP1: AAGTCAGACTACAGGCCAGCCCTGGCTGCC (mutant bases underlined)
FP2: GGCCTGTAGTGTGACTTGAGACCAATAAAAGTGCA (mutant bases underlined)
RP2: AAAAAGCTTGGCCACGCGTCGACTAGTAC (HindIII site underlined) [Note that primer (2) and (3) have overlapping region that includes mutant bases.]
Template and PCR reactions were same as mut1 as described above. The gel purified PCR product was cloned in HindIII and SacI site of pMIR-REPORT™ vector (Ambion), in frame with luciferase and confirmed by sequencing.
Cloning of pre-miRNA
Pre-miRNA sequence of mmu-miR-34b (accession ID MI0000404) was taken from miRBase. The following overlapping primers were used for PCR amplification of pre-miRNA sequence of mmu-mir-34b:
Forward primer: TAATCTAGAGGATCCGTGCTCGGTTTGTAGGCAGTGTAATTAGCTGATTGTAGTGCGGTGC (BamHI recognition sequence underlined)
Reverse primer: ATTGAGCTCAAGCTTGTGCCTTGTTTTTGATGGCAGTGGAGTTAGTGATTGTCAGCACCGCACTACAATCAGCTAATTA (HindIII recognition sequence underlined)
The gel purified PCR product was cloned in HindIII and BamHI site of pSilencer™ 4.1-CMV neo vector (Ambion) and confirmed by sequencing.
Cloning of mmu-miR-34a
The following primers were used for PCR amplification of pre-miRNA sequence of mmu-miR-34a:
Forward primer: AAAGGATCCtgggaCCAGCTGTGAGTAATTCTTTGGCAGTGTCTTAGCTGGTTGTTGTGAGTATTAGC (BamHI sequence underlined, let7 flanking sequence in lower case)
Reverse primer: TTTAAGCTTtaggaACAATGTGCAGCACTTCTAGGGCAGTATACTTGCTGATTGCTTCCTTAGCTAATACTC (HindIII sequence underlined, let7 flanking sequence in lower case)
The gel purified PCR product was cloned in HindIII and BamHI site of pSilencerTM 4.1-CMV neo vector (Ambion) and confirmed by sequencing.
Cloning of mmu-miR-34amut
The following overlapping primers were used for PCR amplification
Forward primer: AAAGGATCCtgggaCCAGCTGTGAGTAATTCTTTGGcACTCTCTTAGCTGGTTGTTGTGAGTATTAGC (BamHI sequence underlined, let7 flanking sequence in lower case, mutant bases are underlined and italics)
Reverse primer: TTTAAGCTTtaggaACAATGTGCAGCACTTCTAGGGCACTCTACTTGCTGATTGCTTCCTTAGCTAATACTC (HindIII sequence underlined, let7 flanking sequence in lower case, mutant bases are underlined and italics)
The gel purified PCR product was cloned in HindIII and BamHI site of pSilencer™ 4.1-CMV neo vector (Ambion) and confirmed by sequencing.
Cell culture and differentiation
Murine neuroblastoma cells (ATCC number CCL-131; Neuro-2a or N2a) were maintained in minimum essential medium (MEM) (GIBCO-BRL) supplemented with 10% fetal calf serum (Biological Industries, Israel), 2 mM l-glutamine (Sigma, USA), 1 mM sodium pyruvate (Sigma) and antibiotic–antimycotic solution (100× stock) (Sigma) at 37°C humidified incubator with 5% CO2. Human embryonic kidney (HEK) 293 T cells were maintained in Dulbecco's modified eagle's medium (DMEM) supplemented with 10% FCS while the rest of the conditions were similar to ones used for the culture of N2a.
Differentiation of Neuro-2a
Approximately 8 × 103 cells were seeded in each well of a two chambered slide (Lab-Tek slide Nunc). Cells were maintained in Opti-MEM (GIBCO-BRL). After 24 h media was replaced by OptiMEM with 5 mM N6,2′-o-dibutyryladenosine-3′,5′-cyclic monophosphate sodium salt (dbcAMP) (Sigma) and incubated up to 4–5 days till differentiation.
In situ RNA localization
Probe preparation
Digoxigenin-UTP labelled RNA probes were prepared by in vitro transcription (IVT) using DIG RNA labelling Kit (T7) (Roche) following manufacturer's protocol. Qiagen column purified PCR products were used as a template for IVT reaction. The following primers were used for PCR amplification of UTR regions that was used in IVT reaction for making sense and antisense probes: T7 promoter sequence has been underlined, in the primer sequences.
Primers used for detection of both short and long UTR:
RNA probe-a
Anti-sense:
FP: CCAACCGTGAAAAGATGACC
RP: TAATACGACTCACTATAGGGTAACAGTCCGCCTAGAAGCA
Sense:
FP: TAATACGACTCACTATAGGGCCAACCGTGAAAAGATGACC
RP: TAACAGTCCGCCTAGAAGCA
Primers used for detection of long UTR:
RNA probe-b
Anti-sense:
FP: GTTTTGGCGCTTTTGACTCAGGATT
RP: TAATACGACTCACTATAGGGGACCAAAGCCTTCATACATCAAGTT
Sense:
FP: TAATACGACTCACTATAGGGGTTTTGGCGCTTTTGACTCAGGATT
RP: GACCAAAGCCTTCATACATCAAGTT
Differentiated cells were fixed with 4% paraformaldehyde for 10 min, washed four times for 5 min with phosphate buffer saline (PBS) supplemented with 0.1% Tween-20 (PBST). Cells were then incubated in pre-hybridization buffer (50% formamide, 5 × SSC, 50 μg ml−1 yeast t-RNA, 50 μg ml−1 heparin, 0.1% Tween-20) for 1 h at 50°C. Digoxigenin-labelled RNA probe was then added and incubated overnight at 50°C. Subsequently, following washes were given: five times with wash1 buffer (2 × SSC, 50% formamide) each with 20 min; one wash with wash2 buffer (2 × SSC, 25 mg ml−1 Rnase A) for 30 min at 37°C; followed by five washes (where SSC buffer was diluted in PBST in a stepwise manner) at room temperature for 15 min. Among those five washes first wash (2× SSC, PBST) was given once, second wash (1× SSC, PBST) and third wash (0.1× SSC, PBST) were performed twice. Cells were kept in blocking solution (2 mg ml−1 BSA, PBST) for 1 h at room temperature and subsequently, incubated with alkaline phosphatase conjugated anti-digoxigenin antibody (Roche) (1 : 300 dilution in PBST) for 30 min at room temperature. After that two washes were given with wash buffer (100 mM Tris–Cl, pH 7.5, 150 mM NaCl) for 15 min at room temperature. Hybridized probes were then detected with NBT/BCIP solution. After a 5-min wash with PBS cells were observed under 60× objective (bright field) in an inverted microscope (NIKON).
Transfection and luciferase assay
Approximately 3 × 105 cells (Neuro 2a) were seeded in each well of a six-well plate (Axygen) 24 h prior to transient transfection when 50–80% confluency was reached. Cells were washed once with Opti-MEM (GIBCO-BRL) and maintained in 1 ml Opti-MEM. Equivalent amount of transfection quality plasmids of vector alone (pMIR-REPORT™), cloned long UTR and short UTR of Actb (Endo free plasmid maxi kit; Qiagen) were used for transfection using Fugene 6 reagent (Roche) following manufacturer's protocol.
Twenty-four hours after transfection, cells were lysed in CCLR buffer (Promega) and total protein was estimated using BCA protein estimation kit (Sigma). Luciferase assay was performed using luciferase assay system (Promega) and luminescence was measured in a microplate luminometer (Berthold detection system). For data analysis, luminescence values were normalized to total protein.
miRNA target prediction and validation experiments
miRNA target prediction was carried out using the miRanda software (http://www.microrna.org/microrna/). We used the long UTR sequence as the input and a cut-off of predicted ΔG ≤ −20 kcal/mol.
Neuro-2a cells were co-transfected with luciferase–long UTR fusion construct and cloned pre-miR-34b. β-Gal plasmid was also co-transfected for normalization control. Luciferase–long UTR in antisense orientation fusion construct/empty vector were used as negative control. Transfection was performed using Amaxa nucleofection method as per manufacturer's protocol.
For LNA experiments approximately 105 Neuro-2a cells per well of a 12-well plate was used for transfection. Neuro-2a cells were co-transfected with luciferase–long UTR fusion construct/correspondingly short UTR construct/empty vector along with antisense LNA oligonucleotide (10−40 nM)/mock LNA oligo (40 nM) and β-gal control plasmid using NeoFX™ (Ambion) transfection reagent following manufacturer's protocol. Due to the high level of sequence similarity the antisense LNA could be able to target both mmu-miR-34b-5p and mmu-miR-34a. The antisense LNA sequences (modified base is underlined) are given below:
ACAACCAGCTAAGA
ACCAATCGACCAAC (Mock LNA or non specific LNA)
Twenty-four hours after transfection cells were lysed in reporter lysis buffer (RLB) (Promega) and reporter assays were performed.
For mutation experiments
Equivalent amount of UTR construct (longUTR/mut1/mut2) was transfected to Neuro-2a cells using 3 μl lipofectamine (Invitrogen) following manufacturer's protocol. β-Gal control plasmid was co-transfected for normalization. Cells (105/well) were plated in 12 well plates before transfection. Twenty-four hours after transfection, cells were harvested for luciferase and β-gal reporter assays.
For compensatory mutagenesis experiments
Approximately 400 ng of long UTR construct/mut2, 400 ng of cloned pre-miR-34a/pre-miR-34amut and 200 ng of β-gal control plasmid were co-transfected into HEK 293T cells using 3 μl lipofectamine (Invitrogen) following manufacturer's protocol. Cells (105/well) were plated in 12 well plate before transfection. Cells were harvested for reporter assays 24 h after transfection.
Reverse transcription followed by PCR
Total RNA was isolated from Neuro-2a cells transfected with luciferase–long UTR fusion construct/luciferase–short UTR construct/empty vector that contains luciferase reporter. RNA isolation method and DNase treatment have been described above. Total RNA was reverse transcribed by using random hexamer and M-MuLV reverse transcriptase (New England Biolabs Inc). cDNA was then PCR amplified by using luciferase-specific primers (FP: CTCGGGTGTAATCAGAAT, RP: TTTACACGAAATTGCTTCT) and 18S rRNA specific primers (FP: CTTTCGAGGCCCTGTAATTG, RP: CCTCCAATGGATCCTCGTTA).
Real time PCR
Real time PCR was performed by using TaqRMan probe specific for mmu-miR-34, miR-92 using TaqRMan microRNA assay kit (Applied Biosystems) following manufacturer's protocol. TaqRMan probes for mmu-miR-34 and miR-92 were purchased from Applied Biosystems. Data were analysed using relative quantification method. miR-92 was used as an endogenous control as per manufacturer's instruction.
Primer extension
Primer extension was performed from 1 μg of total RNA, α-P-32-dCTP, and the following reverse primers:
ACAACCAGCTAAGA (for mmu-miR-34a)
TGCTAATCTTCTCTGTATCG (for mmu-miR-92)
M-MuLV reverse transcriptase (New England Biolabs Inc) was used for primer extension method. Samples were resolved in 20% polyacrylamide gel containing urea (8 M). Radiolabelled gel was scanned in a phosphorimager.
RESULTS
The high-density oligonucleotide array for mouse genome generated by Affymetrix, Mouse U74Av2 includes five probe sets complementary to actin. Three probes (M12481_5_at, M12481_M_at and M12481_3_at) included in this and subsequent versions of the array are complementary to 5′, middle and 3′ parts of the coding region. These probe sets are included on the array as control probes for housekeeping genes and are expected to show high level of signal after hybridization. Two additional probe sets (101578_f_at, 95705_s_at) are also annotated as corresponding to ‘cytoplasmic β-actin gene (Actb)’, but they are complementary to regions 165 bp downstream to the reported 3′-end of the actin transcript (Figure 1). Each probe set consists of overlapping oligonucleotides that cover a part of the Actb gene. The sequences corresponding to the five probe sets are provided in Supplementary Figure 1.
Figure 1.
Mouse cytoplasmic β-actin (Actb) specific antisense hybridization probe sets were mapped on the Actb sequence, a pictorial depiction. M12481_5_at, M12481_M_at, M12481_3_at, 95705_s_at, 101578_f_at are Actb specific antisense probe sets in Affymetrix U74Av2 mouse Genechip. M12481_5_at, M12481_M_at, M12481_3_at are complementary to the coding region of Actb transcript while 95705_s_at, 101578_f_at are complementary to the 3′-UTR region. The nucleotide sequence corresponding to each probe sets are given in Supplementary Figure 1.
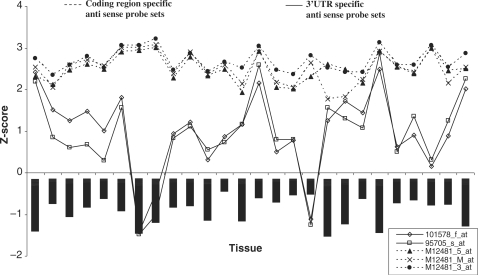
In mouse neuronal gene expression profiling experiments carried out in our lab previously (data not shown), we found that the two probe sets complementary to the Actb downstream region also showed detectable signals. However, the signal was much lower than the signals from the three control probes. This provided an indication that Actb gene may be associated with heterogenous 3′-UTRs. We examined the signals from all five probes in microarray data generated using U74Av2 arrays from Gene Expression Omnibus (GEO), the microarray data repository hosted by NCBI. There are more than 4000 experiments using the GPL81 platform corresponding to the U74Av2 in GEO. We observed that the expression of the three Actb internal probes were consistently higher than the signals from the two downstream probe sets. To explore tissue-specific expression differences between the probes, we used the tissue-specific expression data profiling carried out by Freilich et al. (38). We found that the three internal probes give a consistently high expression level whereas the 3′-UTR-specific probes showed highly variable expression in different tissues (Figure 2). In tissues like pre-putial gland, pyloric andrus and vas deferens, it showed a signal comparable to the high signal of the internal control probe sets, in tissues like appendix, brain and testes medium-level expression was seen whereas extremely low signals were seen in colon and lung. This observation also ruled out the possibility that the 3′-UTR-specific probes simply hybridize less efficiently than the internal probes.
Figure 2.
Tissue-specific expression of cytoplasmic β-actin (Actb). Microarray data (38) was reanalysed and normalized values of Actb specific probes were plotted. Antisense probe sets corresponding to coding region (M12481_5_at, M12481_M_at, M12481_3_at) and 3′-UTR (95705_s_at, 101578_f_at) of Actb are obtained from Affymetrix U74Av2 mouse Genechip.
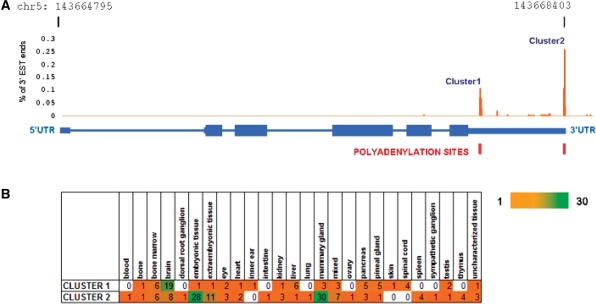
We next carried out EST analysis to validate the results from microarray analysis. ESTs reported from the actin gene locus were systematically analysed to identify longer transcripts which may account for the signal from the downstream probes in the microarrays. The EST analysis revealed two distinct clusters of EST ends in the 3′-UTR of Actb gene, corresponding to the two alternative polyA sites (Figure 3A). The former cluster comprised of a total of 63 ESTs and the latter 122 ESTs. Members of each cluster were further analysed for the tissue of origin using the Unigene annotations (Figure 3B). Brain tissue showed enrichment for the first cluster of ESTs while the latter showed enrichment in the embryonic tissues and mammary gland.
Figure 3.
EST end analysis of Actb transcript revealed two distinct clusters of transcripts corresponding to the two polyA sites (A). The tissue of origin of ESTs in each cluster (B) showed that the ESTs were differentially expressed in different tissues. The scale bar represents abundance of ESTs.
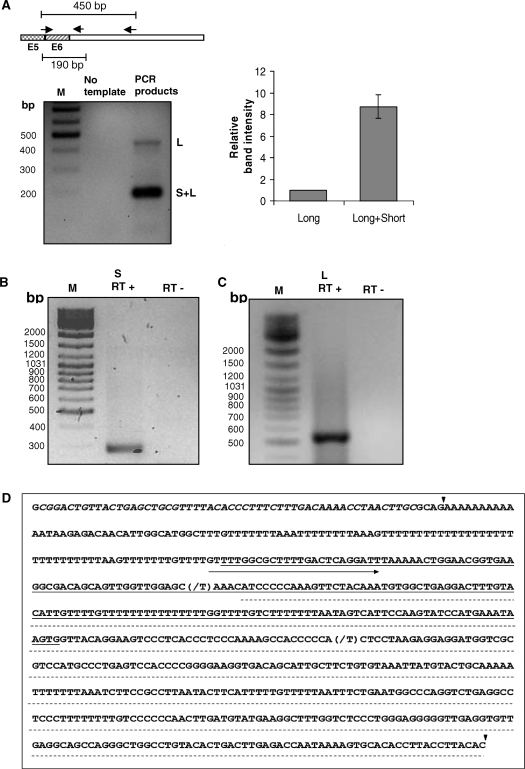
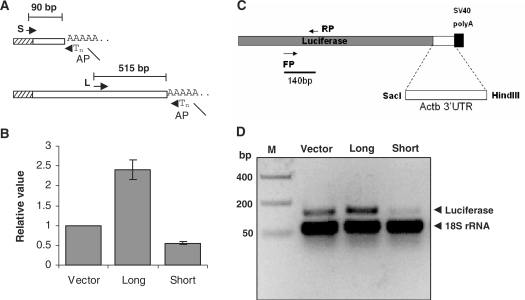
We experimentally verified the presence of alternative 3′-UTRs in cytoplasmic β-actin transcripts. Total RNA isolated from Neuro-2a cells was reverse transcribed to synthesize cDNA and PCR amplification using a common forward primer and alternative 3′-UTR-specific reverse primers revealed the presence of transcripts of varying abundance (Figure 4A). The common forward primer spans the exon junction between the last and penultimate exons. One of the reverse primers is complementary to the region 2 bp following the stop codon and results in a short 190 bp product. This primer can bind to cDNA products from both short and long transcripts. A second reverse primer that binds to the region 259 bp downstream to the stop codon was also included in the analysis. This primer is positioned beyond the predicted proximal polyA site and can bind only to the long transcripts. The 450 bp product arising from this primer reflects the abundance of the long transcript, whereas the abundance of the short 190 bp product represents the collective expression level of short and long transcripts from the Actb gene. These RT–PCR results, in agreement with the microarray and EST analyses showed, without doubt, that mouse neuronal cells express 3′-UTR variants of Actb transcripts. To unambiguously assign the exact 3′-end of the Actb gene, we used the classical 3′ rapid amplification of cDNA ends (RACE) methodology. We carried out 3′-RACE for actin transcripts using total RNA isolated from the mouse neuronal cell line Neuro-2a. The total RNA was used for cDNA synthesis primed by a oligodT primer carrying a 5′ GC-rich AP. The extended region of the AP serves as a reverse primer-binding site for subsequent rounds of PCR amplification using gene-specific forward primers and a common reverse primer. Subsequently, the PCR products were cloned and sequenced. We isolated two distinct types of clones in 3′-RACE experiments that allowed us to unambiguously establish the existence of two alternative transcription stop sites in the Actb gene. The proximal stop site results in a short 3′-UTR of 59 bp that is highly conserved from chicken to human and includes the 54 nt zipcode sequence (Supplementary Figure 2). A gene-specific forward primer positioned 179 bp upstream from the translation stop site, resulted in the ∼290 bp short product (Figure 4B) expected from the proximal polyA site. However, in mouse neuronal cells we also found, additionally, actin transcripts with a much longer 3′-UTR of 678 bp. The gene-specific forward primer was designed to bind a region between the two polyA sites so that it would amplify only the cDNA products of the long transcripts and result in a ∼550 bp product (Figure 4C). The longer transcript includes regions homologous to the two downstream probes (101578_f_at, 95705_s_at) on the Affymetrix array. Figure 4D provides an overview of the sequence elements in the 3′-UTR of the actin transcript along with the positions of the Affymetrix probe sets mentioned here, and the polyA sites identified in our RACE experiments.
Figure 4.
Reverse transcription followed by PCR (RT–PCR) (A) and 3′-RACE (rapid amplification of cDNA 3′-ends) analyses revealed the existence of short (B) and long polyA variant (C) of Actb in mouse neuronal cells. Forward (FP) and reverse (RP) primers (arrow heads) for PCR amplification in RT–PCR analysis were shown by pictorial depiction (A) where FP was an intron spanning primer common for both short and long transcript, E5 and E6: exon 5 and 6, respectively, intronic region between E5 and E6 not shown. Gene-specific FP and AP for 3′-RACE analysis were described in Materials and methods section. The intensity of each band in A, left panel was quantified and plotted (A, right panel) relative to long UTR-specific band. Data represent mean ± SD of two independent experiments performed. (D) 3′-RACE products were sequenced and first and second cleavage sites (filled inverted triangle), FP used in RACE for amplifying long UTR (right arrow), Affymetrix probe sets 95705_s_at (underlined) and 101578_f_at (dashed line), Zip code (italics) demarcated on the 3′-UTR sequence. Nucleotide that differs from Genebank sequence was indicated as (slash). M: DNA marker, S and L: transcript bearing short 3′-UTR and long 3′-UTR, respectively. RT+: with reverse transcriptase, RT−: without reverse transcriptase.
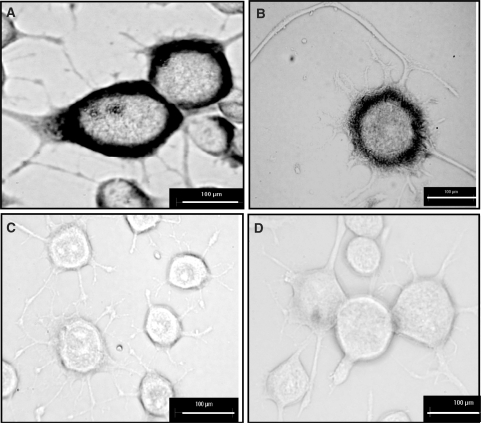
Our RT–PCR analysis implied that the longer transcripts were less abundant than the short transcript. However, to rule out the possibility that the PCR product abundance may be affected by the efficiency of PCR, we carried out in vivo expression analysis. We performed in situ hybridization using probes specific to the long UTR and common to both transcripts in neuronal cells to look for differential localization of the longer actin transcript. As shown in Figure 5, both long transcript specific antisense probe and the common antisense probe showed cytoplasmic localization while sense probes failed to show any signal. The signal from the common antisense probe that can bind to both long and short transcripts was consistently higher than the signal from the long transcript specific probe. Therefore, in keeping with the microarray data, the longer transcript seems to be less abundant in neuronal cells.
Figure 5.
Existence of polyA variants of Actb in differentiated mouse neuronal cell analysed by in situ hybridization. Differentiated Neuro-2a cells were hybridized with digoxigenin-UTP labelled antisense RNA probe-a (A), sense probe-a (C), antisense RNA probe-b (B), sense probe-b (D). Antisense RNA probe-a: antisense probe that can detect both short and long polyA variants; antisense RNA probe-b: antisense probe specific for long polyA variant (scale bar, 100 μm).
We cloned the short and the long UTR regions (Figure 6A) isolated in our RACE experiments downstream of the luciferase reporter gene driven by the highly and constitutively expressed CMV promoter. The luciferase activity per unit total protein showed that the long 3′-UTR region conferred 2.4-fold higher steady state expression level of the reporter as compared to the vector transfected controls (Figure 6B). The expression level of the reporter from the short UTR-reporter fusion construct was reduced to nearly half of the vector control. The relative difference in luciferase activity between the short and long 3′-UTR clones was 4.3-fold. The difference in reporter activity seems to arise from differential transcript stability since the luciferase transcript levels normalized to 18S rRNA levels, in RT PCR experiments, showed relatively higher levels of the long UTR transfected cells (Figure 6D). The additional 515 bp in the longer transcript seems to harbour sequences that stabilize the transcript or improve translational efficiency, since protein levels from the reporter gene are significantly higher in the cells expressing the long transcript fused to the reporter gene.
Figure 6.
Effect of short and long 3′-UTR sequence on translation. (A) Sequence region of short and long 3′-UTR of Actb was cloned (see Materials and Methods section) in a pmirReport vector (Ambion) containing luciferase as a reporter. Cloned region of short- and long-UTR were pictorially described (A) by primer sets where reverse primer was AP used for RACE; short- and long-UTR-specific forward primers were denoted by S and L, respectively, followed by arrow heads. (B) Translational effect of UTR sequences were determined by luciferase assay, 24 h after transfection cells were lysed and luminescence were counted in a luminometer. Data were normalized to total protein and represented relative to vector transfected control. Data shown are mean ± SEM of four independent experiments, in replicate performed. (C, D) Effects of UTR sequences on mRNA level were checked by RT–PCR (D). Total RNA was isolated from cells 24 h after transfection and reverse transcribed, further luciferase (reporter) and 18S rRNA specific primers were used for PCR to check mRNA level and loading control, respectively. Luciferase specific primers were shown (C).
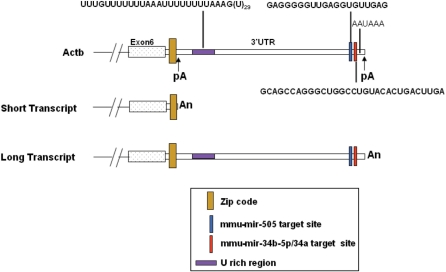
We next looked for potential microRNA target sites in the 3′-UTR region of Actb. Using miRanda, we predicted mmu-miR-34b-5p and mmu-miR-505 binding sites (Figure 7) in the long UTR of Actb transcript, downstream to the proximal polyA sites. Figure 7 provides a schematic representation of the sequence features identified in the actin 3′-UTR sequence, including localization signals, miRNA binding sites experimentally identified polyA sites and associated sequence elements. On closer inspection, it was seen that the mmu-miR-34a and mmu-miR-34b-5p had identical seed sequences and therefore both could potentially target the same site. We checked the evolutionary conservation at these sites, since functional miRNA target sites in the 3′-UTR are likely to show higher conservation than regions that do not harbour functional elements. The mmu-miR-34b-5p/34a target site was highly conserved amongst vertebrates (Supplementary Figure 3A), while the mmu-miR505 site is deleted in the human homologue (Supplementary Figure 3B). We also saw that in Drosophila, dre-miR-34, shows a potential binding site in the actin transcript (data not shown). Due to poor conservation of the target site, mmu-miR-505 was not validated further. In summary, the longer transcript arising from the distal polyA site seems to be less abundant, translationally more efficient and potentially targeted by mmu-miR-34b-5p/34a.
Figure 7.
Annotation of 3′-UTR of mouse cytoplasmic β-actin (Actb) reference sequence along with short and long polyA variants. pA: polyA site.
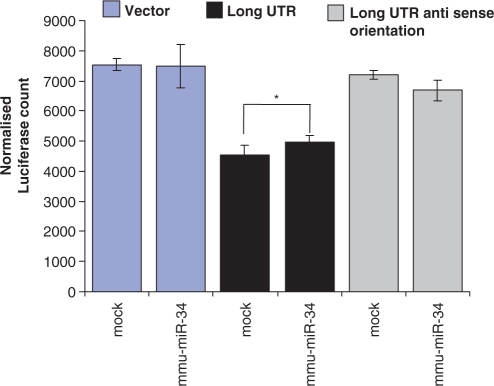
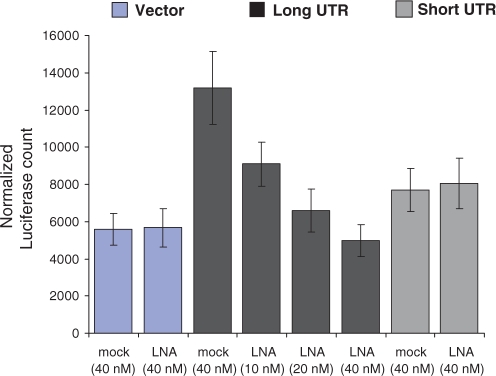
We attempted to study the role of miRNA-mediated regulation on the long UTR of Actb by over expressing the pre-miRNA. We expressed pre-miR-34b under the CMV promoter in Neuro-2a cells along with the luciferase–UTR fused constructs. Cells expressing the miRNA and long UTR construct showed a modest up-regulation (statistically significant P-value <0.05, Student's t-test) of luciferase reporter. The long UTR in the antisense orientation was not affected (P-value >0.05, Student's t-test) as compared to mock (pSilencer negative control vector) transfected control (Figure 8). Next, we used anti-miRNA molecules to study the effect of down-regulation of endogenous mmu-miR-34a/34b-5p. The high sequence similarity between miR-34a/34b-5p sequences allows the anti-miRNA molecule to target both mmu-miR-34a and mmu-miR-34b-5p. We have earlier compared unmodified and locked nucleic acid (LNA) modified oligonucleotides for this purpose and found that LNA modification results in higher affinity for the target miRNA sequence and interference at lower concentration minimizing non-specific and toxic effects (unpublished result). Anti-miRNA molecules resulted in up to 3-fold down-regulation of expression from long UTR whereas short UTR remained unaffected (Figure 9). Endogenous mmu-miR-34a/34b-5p, therefore, mediates higher expression from the long UTR.
Figure 8.
mmu-miR-34 dependent regulation of long 3′-UTR variant of Actb in Neuro-2a cells. Long UTR-specific region was cloned into pmirReport vector (Ambion) that contains luciferase as a reporter under CMV promoter. Long UTR was cloned in anti-sense orientation into the same vector. Pre-miRNA of mmu-miR-34b was cloned into pSilencer 4.1-CMV neo vector (Ambion). Neuro-2a cells were co-transfected with long UTR construct/long UTR anti-sense orientation construct/empty vector along with pre-miR-34b construct/mock. pSilencer vector (Ambion) was used as negative control (mock). Twenty-four hours after transfection cells were harvested for assays. Reporter (luciferase) activity was normalized to the activity of a co-transfected CMV-β-galactosidase construct. Data represented the mean ± SEM of two independent experiments, each in duplicate, performed. (*P < 0.05, Student's t-test).
Figure 9.
Inhibition of mmu-miR-34a/b by transfection of antisense LNA oligonucleotide leads to the down-regulation of long 3′-UTR variant of Actb in a concentration dependent manner. Antisense LNA oligonucleotide was designed against both mmu-miR-34b-5p/34a; and co-transfected with UTR constructs of Actb and empty vector into Neuro-2a cells. A non-specific LNA was used as a negative control (mock). Twenty-four after transfection cells were harvested for assays. Reporter (luciferase) activity was normalized to the activity of a co-transfected CMV-β-galactosidase construct. Data represented the mean ± SEM of two independent experiments, each in duplicate, performed.
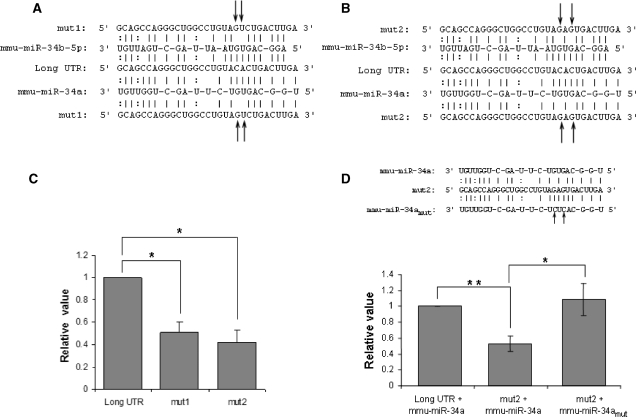
To study the effect of mmu-miR-34a/34b-5p in further details, we created two mutants constructs each carrying two mutations in the miRNA target region (Figure 10A and B). The mut1 construct carried two mutations in the target region, at adjacent positions in the middle of the seed region. Mutations in the middle of the seed region are expected to result in maximum destabilization of the miRNA target binding. The mut2 construct also carried two mutations disrupting two G-C bonds in the miRNA target binding. As shown in Figure 10C, both 3′-UTR mutations resulted in reduced expression of the luciferase reporter confirming that the miRNA target region mediates high expression of the long UTR of the mouse Actb gene in neuronal cells. The mut2 construct showed marginally higher effect resulting in nearly 60% reduction in expression compared with the long UTR construct. We next created constructs bearing mutant pre-mmu-miR-34a with mutations that would restore binding to mut2 construct. We incorporated five bases from the let-7 miRNA flanking regions adjacent to the pre-miRNA to ensure over expression of the miRNA. The expression of the mature miRNA was verified by quantitative real time PCR (Supplementary Figure 4). In cells over-expressing wild-type mmu-miR-34a, consistent with Figure 10C, the mut2 construct showed lower expression levels compared to the long UTR. The expression of the reporter was restored when the mut2 construct was expressed with the mutant mmu-miR-34a bearing compensating mutations that allow miRNA target binding (Figure 10D). Therefore, mmu-miR-34a mediates up-regulation of the long UTR of the Actb gene.
Figure 10.
Mutation in the miRNA (mmu-miR-34a/b-5p) binding site of long 3′-UTR of Actb affects translation. (A and B) Mutant versions (named mut1 and mut2) of long 3′-UTR of Actb (long UTR) were prepared by introducing two base changes (indicated by arrows) to the miRNA (mmu-miR-34a/b-5p) seed paring region. (C) Translational effect of mutation in the miRNA seed pairing region of long UTR was analysed by reporter (luciferase) assay. Neuro-2a cells transfected with long UTR/mut1/mut2 were harvested for assay after 24 h of transfection. Reporter (luciferase) activity was normalized to the activity of a co-transfected CMV-β-galactosidase construct. Normalized luciferase activity was represented relative to the long UTR transfected control. Data shown are mean ± SEM of two independent experiments performed. (*P < 0.05, Student's t-test). (D) Mutations in the seed region of mmu-miR-34a compensate for mutation in the target region of long UTR; indicates a direct effect of mmu-miR-34a on Actb long UTR. Mutant version of mmu-miR-34a (named mmu-miR-34amut) was prepared by introducing two base changes (indicated by arrows) complementary to the mutations in mutant version of long UTR (mut2). HEK 293 T cells were co-transfected with long UTR/mut2 along with cloned mmu-miR-34a/mmu-miR-34amut and cells were harvested for assays after 24 h of transfection. Reporter (luciferase) activity was normalized to the activity of a co-transfected CMV-β-galactosidase construct. Normalized luciferase activity was represented relative to the long UTR transfected control. Data shown are mean ± SEM of three independent experiments performed (*P < 0.05, **P < 0.01, Student's t-test).
DISCUSSION
Recent bioinformatics studies have shown that nearly 50% of genes in the human genome may harbour alternative polyA sites (33). Heterogeneity in 3′-UTRs can therefore account for differential regulation, stability and spatio-temporal expression of transcripts from the same gene. We have used EST and gene expression data, 3′-RACE analysis and in situ hybridization to establish that the cytoplasmic β-actin gene generates two alternative transcripts terminated at tandem polyA sites. Our analysis indicates that the longer transcripts are regulated in a tissue-specific manner, whereas the proximal polyA site is used for constitutive expression. Functionally, the longer UTR confers higher stability on the transcript, resulting in higher reporter gene expression, in mouse neuronal cells. Besides its highly conserved housekeeping function, the Actb gene plays diverse roles. It has been suggested that variable 3′-UTRs may contribute to the complexity of transcriptome and provide a mechanism for differential regulation in a tissue-specific manner (37). The existence of tandem polyA site and heterogeneous 3′-UTRs in the well-studied Actb gene provides experimental support for the involvement of 3′-UTR in tissue-specific gene expression.
We explored the sequences surrounding the polyA site for conserved motifs that have been shown to flank the cleavage site. The polyA is carried out by the polyA polymerase in mammalian cells in a highly regulated manner, through a complicated mechanism involving multi-protein complexes that interact with these sequences on either side of the cleavage site. The most well-characterized sequence element that signals polyA sites is the AAUAAA motif found within 10–30 nt upstream of the cleavage site (26,27). The distal polyA site is associated with a canonical PAS element followed by downstream G/U rich sequences and has been bioinformatically identified in whole genome studies. We did not find a canonical polyA element upstream of the short transcript. The AAUAAA sequence can account for only about 53% of all the known polyA sequences in the genome (28). The major sequence variant of the PAS element, AUUAAA, accounts for an additional 16% of all polyA sites predicted by EST mapping (28). Several genes harbour other variants of the polyA site, although they vary in strength and maybe enhanced by the presence G/U or U rich sequences downstream. Nearly 13 most frequently occurring variants of the PAS site have been identified by large-scale analysis of polyA sites in the human and mouse genomes (28,29). Although the canonical PAS element is not seen with 40 nt upstream of the proximal polyA site, we found a UUUACA hexamer at 31 nt upstream of the proximal site. This hexamer has a single substitution at the fifth position from the hexamer UUUAAA which is amongst the 10 most frequently found hexamers reported in previous studies. There is a strong U rich region 35 nt downstream of the proximal cleavage site that may enhance use of the non-canonical upstream signal.
EST and microarray analysis indicated that the long transcript is highly expressed in embryonic and germinal tissue, since detection levels were high in pre-putial gland, vas deferens and testes (microarray analysis) and embryonic tissue and mammary gland (EST analysis). The whole brain showed a medium level of expression of the long transcript in microarray and EST data whereas it was poorly expressed in other tissues. The inclusion of several cell types in whole brain samples and the resultant heterogeneous nature of whole brain RNA can result in dilution of expression level of transcripts that maybe expressed in a specific subpopulation of neuronal cells. The actin transcript has been shown to undergo rapid deadenylation during early developmental stages in mouse embryos (8,9). In neuronal cells, the actin transcript is localized to the synapses as part of ribo-nucleoprotein bundles (40). Transient localization of actin transcripts following exposure to neutrophins and brain-derived neurotrophic factor (BDNF) indicates that it has an important step in directed growth of axons and synaptic connectivity (41). The localization of actin transcript requires a sequence element called the zipcode (7). Reporter β-galactosidase fusion to the zipcode results in localization of lacZ expression to sites of actin polymerization at the periphery of chick fibroblast cells (7). The zipcode is a conserved 54 nucleotide long sequence that occurs in chick, dog, mouse and human actin transcripts, immediately following the stop codon. Both the short and long UTRs identified in our study, contain the zipcode sequence. The heterogeneity in the UTR apparently does not disrupt the localization signal. However, the structural basis of the binding of the zipcode binding factor (ZBF) to the localization signal has not been studied. The additional regions in the longer transcript could potentially alter the secondary structure of the transcript and modulate the localization of actin mRNA to the cell periphery. Our in situ hybridization results serve to demonstrate the in vivo presence of the longer transcript in neuronal cells and provide a basis for studying specific localization of these transcripts to growth cones of axons following treatment with external agents like neutrophins and BDNF.
The longer UTR harbours target sites for regulation by small regulatory RNAs. MicroRNAs are 21–23 nt long regulatory RNAs that can bind to target sites of partial complimentarity within UTRs and interfere with transcript stability or translational efficiency. Alternative polyA at tandem sites can result in transcripts coding for the same protein with differential susceptibility to miRNA-mediated regulation at the translational level. We found that the longer actin transcript harbours potential binding sites for mmu-miR-34a/34b-5p and mmu-miR 505. The human, mouse and rat transcripts show high level of conservation at the miR-34a/34b-5p binding sites, implying that these sites are functional. We observed a pronounced dose dependent down-regulation of luciferase reporter activity in cells transfected with LNA modified anti-miR-34a/34b-5p oligonucleotides and luciferase–long UTR fusion construct. Moreover, compensatory mutagenesis of the miRNA and target conclusively showed the positive effect of the miRNA on target expression. Taken together, these results allow us to attribute a mmu-miR-34b-5p/34a mediated positive effect on expression from the long UTR of Actb. miRNAs are generally believed to have a repressive effect on target molecules. Although rare, miRNAs have earlier been shown to mediate a positive regulatory effect in specialized conditions. For instance, the hsa-miR-369-3p binds to AU rich elements (ARE), in the 3′-UTR of tumour necrosis factor-α (TNF-α) transcript, recruits specific proteins and activates translation under growth arrest conditions (42). A similar miRNA-mediated up-regulation of target transcript has been demonstrated by Let-7 at the 3′-UTR of high mobility group A2 (HMGA2) (42). mmu-miR-34a/34b-5p mediated up-regulation of Actb long UTR transcript is part of a growing body of evidence that miRNA can mediate repressive as well as inducing effect on gene expression.
In lower organisms like Drosophila, the miR-34 family consists of a single miRNA whereas in human, it is represented by highly similar sequence variants hsa-miR-34a, b and c (43). The human miRNA, hsa-miR-34a can be induced by ionizing radiation and DNA damage agents in a p53-dependent manner (44,45). It has been shown to be highly expressed in pancreatic cancers, actively participate in the p53 tumour suppressor pathway (46,47). Its targets include Notch, Delta1 and the transcription factor E2F3, besides the anti-apoptotic gene bcl2 (48,49). The existence of mmu-miR-34a/34b-5p target sites in the long UTR of Actb implies that further experimental analysis of differential expression of the 3′-UTRs in cancers can be important. The actin transcript has been found in ribonucleoprotein complexes in neuronal synapses, but miR-34 was not found to be abundant in cell neurites, in the first study to identify miRNAs localized to the neurites (50). Further studies of co-localization of the mRNA and miRNA in axons and dendrites of specific neuronal cells may reveal the functional relevance of the 3′-UTR heterogeneity in Actb transcripts.
In summary, our studies unambiguously establish the presence of tandem alternative polyA sites and 3′-UTRs of different tissue-specific expression pattern in Actb mRNA transcripts. This is the first report of heterogeneity in transcripts of the Actb gene, widely held to be a constitutively and ubiquitously expressed housekeeping gene. It supports the findings of several bioinformatics studies that uncovered the extensive occurrence of polyA sites in the genome. Furthermore, this study illustrates that the integrated analysis of microarray data, EST data and miRNA target prediction can provide novel insights into differential regulation and functional importance of alternative polyA in the genome. The approach used here, of using differential hybridization signals from different probe sets to identify alternative splicing and polyA can be used further for genome-wide bioinformatics analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Council of Scientific and Industrial Research (CSIR) (Project title: Comparative Genomics and Biology of non-coding RNA), India. Funding for Open Access charges: CSIR, India.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sridhar Sivasubbu for advice in RACE experiments and S. Maiti for carefully reviewing the manuscript. T.G. is a recipient of senior research fellowship from CSIR, India.
REFERENCES
- 1.Miralles F, Visa N. Actin in transcription and transcription regulation. Curr. Opin. Cell Biol. 2006;18:261–266. doi: 10.1016/j.ceb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat. Rev. Mol. Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 3.Hu P, Wu S, Hernandez N. A role for beta-actin in RNA polymerase III transcription. Genes Dev. 2004;18:3010–3015. doi: 10.1101/gad.1250804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 2004;6:1094–1101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 5.Grummt I. Actin and myosin as transcription factors. Curr. Opin. Genet. Dev. 2006;16:191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Ponte P, Ng SY, Engel J, Gunning P, Kedes L. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. Nucleic Acids Res. 1984;12:1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachvarova R, De LV, Johnson A, Kaplan G, Paynton BV. Changes in total RNA, polyadenylated RNA, and actin mRNA during meiotic maturation of mouse oocytes. Dev. Biol. 1985;108:325–331. doi: 10.1016/0012-1606(85)90036-3. [DOI] [PubMed] [Google Scholar]
- 9.Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev. Biol. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- 10.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu. Rev. Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 11.Keller W. No end yet to messenger RNA 3′ processing! Cell. 1995;81:829–832. doi: 10.1016/0092-8674(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 12.Manley JL. A complex protein assembly catalyzes polyadenylation of mRNA precursors. Curr. Opin. Genet. Dev. 1995;5:222–228. doi: 10.1016/0959-437x(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 13.Proudfoot N. Ending the message is not so simple. Cell. 1996;87:779–781. doi: 10.1016/s0092-8674(00)81982-0. [DOI] [PubMed] [Google Scholar]
- 14.Gilmartin GM. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev. 2005;19:2517–2521. doi: 10.1101/gad.1378105. [DOI] [PubMed] [Google Scholar]
- 15.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford LP, Bagga PS, Wilusz J. The poly(A) tail inhibits the assembly of a 3′-to-5′ exonuclease in an in vitro RNA stability system. Mol. Cell Biol. 1997;17:398–406. doi: 10.1128/mcb.17.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtis D, Lehmann R, Zamore PD. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 20.Jackson RJ, Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 21.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 22.Preiss T, Muckenthaler M, Hentze MW. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheets MD, Fox CA, Hunt T, Vande WG, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 24.Eckner R, Ellmeier W, Birnstiel ML. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, Carmichael GG. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proudfoot N. Poly(A) signals. Cell. 1991;64:671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- 27.Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 28.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 31.Tabaska JE, Zhang MQ. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- 32.Legendre M, Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwalds-Gilbert G, Veraldi KL, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin B, Rommens JM, Graham RK, Kalchman M, MacDonald H, Nasir J, Delaney A, Goldberg YP, Hayden MR. Differential 3′ polyadenylation of the Huntington disease gene results in two mRNA species with variable tissue expression. Hum. Mol. Genet. 1993;2:1541–1545. doi: 10.1093/hmg/2.10.1541. [DOI] [PubMed] [Google Scholar]
- 36.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freilich S, Massingham T, Bhattacharyya S, Ponsting H, Lyons PA, Freeman TC, Thornton JM. Relationship between the tissue-specificity of mouse gene expression and the evolutionary origin and function of the proteins. Genome Biol. 2005;6:R56. doi: 10.1186/gb-2005-6-7-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J. Neurosci. 2003;23:3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths-Jones S, Grocock RJ, van DS, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 45.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 46.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 47.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 49.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl Acad. Sci. U.S.A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.