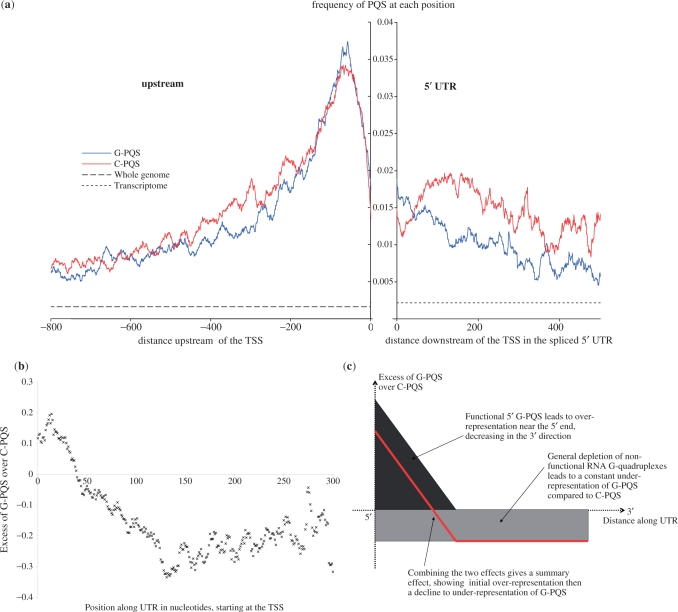
Figure 3.
(a) High-resolution map of PQS at the TSS junction. Distances are shown in units of bases in the 5′ direction (−ve) or 3′ direction (+ve). The frequencies represent the number of times a PQS was observed to include each individual base position, normalized for the number of times each position was observed. The equivalent frequencies across the whole genome and transcriptome are also shown for comparison. The upstream region is taken from whole genomic data, the 5′-UTR data is taken from the mature mRNA post-splicing, with a gap to highlight the junction. (b) Analysis of the excess of G-PQS over C-PQS for 5′-UTRs in the human genome, calculating the excess of G-PQS over C-PQS as Excess = (G-PQS − C-PQS)/(G-PQS + C-PQS) at every position along the 5′-UTR, counting in bases from the TSS. (c) A simple model to predict over-/under-representation of G-PQS motifs compared to C-PQS motifs suggests an overall profile given by the red line, in good agreement with the observed data.