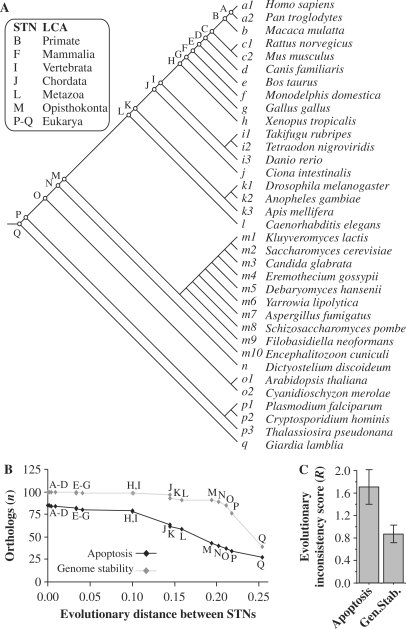
Figure 2.
Inferring evolutionary roots of human apoptosis and genome-stability genes. (A) Eukaryote species tree topology used in the parsimony analysis. The phylogenic relationship among these 35 eukaryotes is based on a manual integration of a variety of phylogenies (28–33). STNs and the corresponding LCA are indicated. (B) Distribution of apoptotic and of genome-stability orthologs according to the roots inferred in the species tree and plotted as a function of the divergence between STNs (based on branch-length estimates). In Supplementary Material Online we exemplified the parsimony analysis. The evolutionary distances were computed using three protein families regarded as very conserved among distant taxa and described as able to reconstruct the three-domain phylogeny: 40S ribosomal proteins, translation initiation factor 5A proteins and Flap structure-specific endonuclease 1 proteins (73). All proteins used in the analysis are aligned in Supplementary Figures S10–S12. The distances are expressed as the fraction of sites that differ between the branches in a multiple alignment, which is an approximation of the branch-length that separates STNs. (C) Divergence between KOG and Inparanoid-derived scenarios. For apoptosis genes, R = 1.709 STNs ±0.224 (SE) and for genome-stability genes R = 0.807 STNs ±0.202 (SE). It means that for each root inferred in our analyses, the estimated error for apoptosis is approximately two STNs up and down from the rooting point in the species tree, while for genome stability the error is approximately one STN up and down.