FIG. 2.
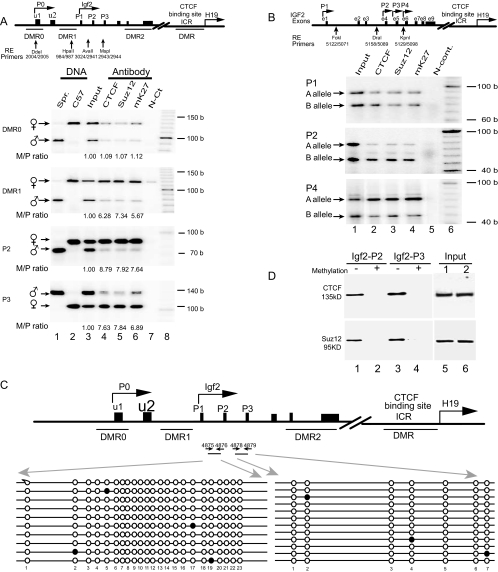
Allele-specific ChIP assay across DMR0, DMR1, and the promoter region of Igf2. (A, top) Scheme of the Igf2/H19 imprinting domain. The exons are depicted as solid boxes. DMRs are shown as underlines, and the polymorphic restriction enzyme sites are shown as vertical arrows. The allelic interaction of CTCF with the Igf2 promoters (P0 to P3) and DMRs was identified by using four polymorphic restriction enzymes (Dde1, Hpa2, Ava1, and Msp1) that distinguish M. spretus from C57BL/c. (A, bottom) ChIP of CTCF, Suz12, and dimethylated H3-K27 (mK27) in F1 mouse fibroblasts derived from breeding M. spretus males with C57BL/c females. Cross-linked DNA-protein complexes were immunoprecipitated with antisera against CTCF, Suz12, and dimethyl-H3-K27 (mK27), followed by PCR amplification with specific primers for the DMR0, DMR1, and Igf2 promoters (P1 to P3). Allelic ChIP products were distinguished by polymorphic restriction enzymes (RE). N-Ct lane, negative control (no antibody); input lane, genomic DNA collected before antibody precipitation (positive control). The M/P ratio is the ratio of the maternal to the paternal alleles after normalization with the input DNA. (B) ChIP of CTCF, Suz12, and dimethylated H3-K27 in human fibroblasts. Alleles are labeled A and B because the parental alleles are not known. (C) DNA methylation of CpG dinucleotides at the Igf2 promoters. After sodium bisulfite treatment, genomic DNA fragments were amplified with primers 4875 and 4876 for promoter P2 and primers 4878 and 4879 for promoter P3. Each line represents a single sequenced PCR molecule. Black circles represent methylated CpG dinucleotides, and open circles represent unmethylated CpG dinucleotides. Note the DNA hypomethylation in both promoters. (D) Oligonucleotide pull-down assay for CTCF and Suz12 with Igf2 promoter P2 and P3 DNA fragments labeled with biotin-streptavidin. Wild-type (−) and methylated (+) DNA fragments were end labeled with biotin and incubated with nuclear extracts. After incubation, DNA fragments were pulled down with streptavidin beads. Proteins bound to the DNA fragments were eluted and detected by Western blotting with antibodies directed against CTCF and Suz12. (Input) Aliquots of nuclear proteins, collected before oligonucleotide pull down, were analyzed in parallel with the samples in lanes 1 to 4 and detected by Western blotting. b, bases.