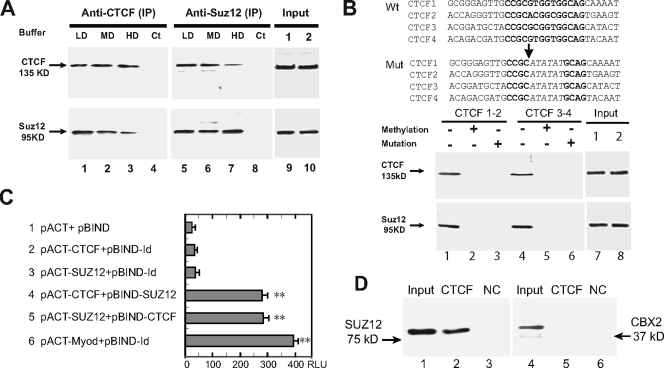
FIG. 3.
(A) Interaction between CTCF and Suz12 as measured by co-IP and Western immunoblot assays. Nuclear proteins were immunoprecipitated, respectively, with anti-CTCF and anti-Suz12 antibodies and washed with detergent buffers. After separation on a 7.5% SDS-polyacrylamide gel, immunoprecipitates were immunodetected with appropriate antibodies to CTCF and Suz12. LD, low-detergent buffer; MD, medium-detergent buffer; HD, high-detergent buffer; Ct, negative IP control with no antibody. (Input) Aliquots of nuclear proteins, collected before CTCF and Suz12 IP, were analyzed in parallel with the samples in lanes 1 to 8 and detected by Western blotting. (B) Oligonucleotide pull down of CTCF and Suz12. At the top are the sequences of the wild-type (Wt) and mutated (Mut) CTCF-binding sites in synthesized oligonucleotide fragments. Consensus CTCF-binding sites are in bold and are partially replaced by ATATAT in mutated oligonucleotides. (Input) Aliquots of nuclear proteins, collected before CTCF oligonucleotide pull down, were analyzed in parallel with the samples in lanes 1 to 6 and detected by Western blotting. At the bottom is Western blotting of CTCF and Suz12 in nuclear proteins pulled down by wild-type (−), methylated (+), and mutated (+) CTCF oligonucleotide fragments. DNA fragments were end labeled with biotin and incubated with nuclear extracts. After incubation, DNA fragments were pulled down with streptavidin beads. Proteins bound to the DNA fragments were eluted and detected with CTCF and Suz12 antibodies. (C) Two-hybrid interactions between CTCF and Suz12. Lane 1 shows background expression of firefly luciferase from the pG5luc vector as determined by cotransfection with the pACT and pBIND vectors, which did not contain CTCF or Suz12, into HBF1 human fibroblast cells. Lanes 2 and 3 show two controls used to determine the background activity of individual CTCF or Suz12 (no interaction). In lanes 4 and 5, The reporter vector was cotransfected with CTCF and SUZ12 in fusion with the VP16 transcription activation domain (pACT constructs) or the GAL4 DNA-binding domain (pBIND constructs). In lane 6, pBIND-Id and pACT-MyoD were used as the positive control encoding two proteins known to interact in vivo. Luciferase activity was measured as relative luminescence units (RLU). *, P < 0.01 compared with the three control groups. The values shown are averages ± standard deviations (n = 6). (D) In vitro binding assay with recombinant proteins. Lanes 1 to 3 show the CTCF-and-Suz12 interaction; lanes 4 to 6 show the CTCF-and-CBX2 interaction. Input, reaction mixture aliquot collected before particle pull down and analyzed in parallel with the samples (CTCF-GST and BSA) by Western blotting. NC, negative control with an equal amount of BSA.