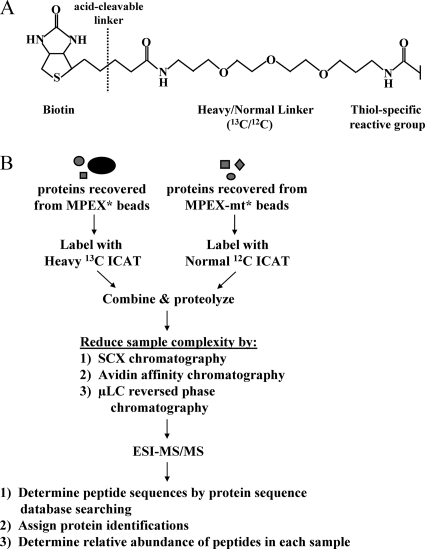
FIG. 3.
Quantitative proteomic identification of MPEX-BF candidates. (A) Structure of the ICAT tag. (B) Quantitative proteomic strategy for identifying MPEX-BFs. Protein samples recovered from MPEX* and MPEX-mt* beads were labeled with isotopically heavy (13C) or normal (12C) forms of the ICAT reagent, respectively. The labeled protein samples were combined and proteolyzed with endoproteinase Lys-C and trypsin. Sample complexity was then reduced by sequential chromatography. Peptides were fractionated by SCX chromatography, after which ICAT-labeled, cysteine-containing peptides were isolated from each of six fractions by avidin affinity chromatography. Peptides in each fraction were further resolved and analyzed by μLC-electrospray ionization (ESI)-MS/MS. Peptide sequences were determined by searching MS/MS spectra against a mouse protein database using the search algorithm SEQUEST. The relative abundance of peptides recovered from MPEX* versus MPEX-mt* beads was determined from the ratio of the heavy and normal signal intensities for each peptide pair. Heavy/normal ratios of at least 2:1 indicate selective enrichment of the corresponding candidate protein within the nuclear factor complex bound to oligonucleotides containing the wild-type MPEX sequence.