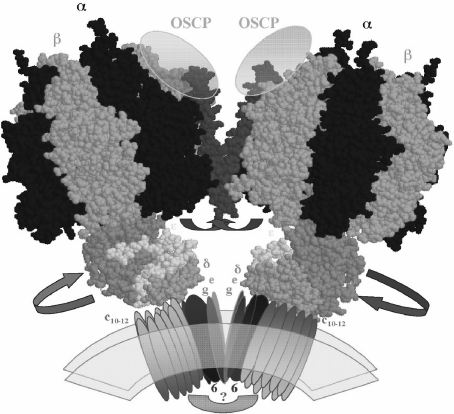
Fig. 4.
Model of the dimeric-mitochondrial ATP synthase: possible localization of the IF1 protein and its movements to allow rotation of the central stalk during ATP synthesis. The model depicts the overall shape of the dimeric ATP synthase molecule that we observed for the bovine mitochondrial enzyme [54]. The dimeric interface involves F0 subunits (e and g) and two protein bridges, one at the F0–F0 side of unknown composition (question mark) and another at the F1–F1 interface where the second stalks (not shown for clarity) and the IF1 protein (red) are likely to be located. The C-terminal side of the IF1 molecule is assumed to cross the dimer interface and to stabilize the dimer by interacting with subunits OSCP [65] and possibly subunits of the second stalk. The N-terminal inhibitory domain that in the absence of the proton gradient blocks rotation of the central stalk by entering at an α–β–γ interface (Fig. 2) is removed from this position and exposed into the media after establishment of a transmembrane proton gradient, thus allowing rotation of the central stalk during ATP synthesis. The F1 structures were constructed from the bovine F1-DCCD coordinates available (PDF code 1E79)