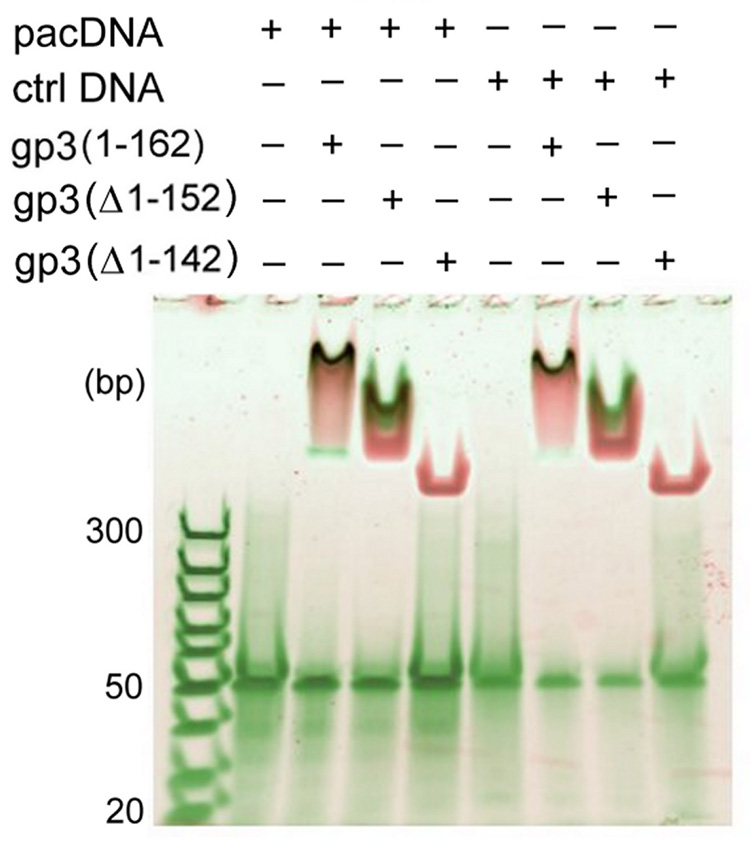
Figure 1.
The binding of full-length and C-terminally truncated gp3 rings to 50-bp DNA targets incorporating either the 21-bp pac site or a permuted (nonfunctional) sequence of the same AT/GC content26 was examined by native gel electrophoretic mobility shift assay. Mixtures of DNA and gp3 rings were incubated at room temperature for 30 min and loaded onto a 4–20% polyacrylamide gel. The total DNA concentration in each lane was 200 nM. DNA and protein were stained with SYBR Green and SYPRO Ruby fluorescent dyes, respectively, and the gel was scanned with a Typhoon 9400 Scanner (Amersham Biosciences, Piscataway, NJ). The DNA-free gp3 ring appears at the top of the gel and protein-free DNA (50 bp) migrates to the middle of the gel. DNA/gp3 complexes are visualized as dark spots. The full-length and truncated gp3 rings progress differently into the gel due to the different net charges of the respective rings. While rings assembled from full-length gp3 and the gp3(Δ1–152) truncate bind DNA comparably, no DNA binding is observed for the gp3(Δ1–142) ring.