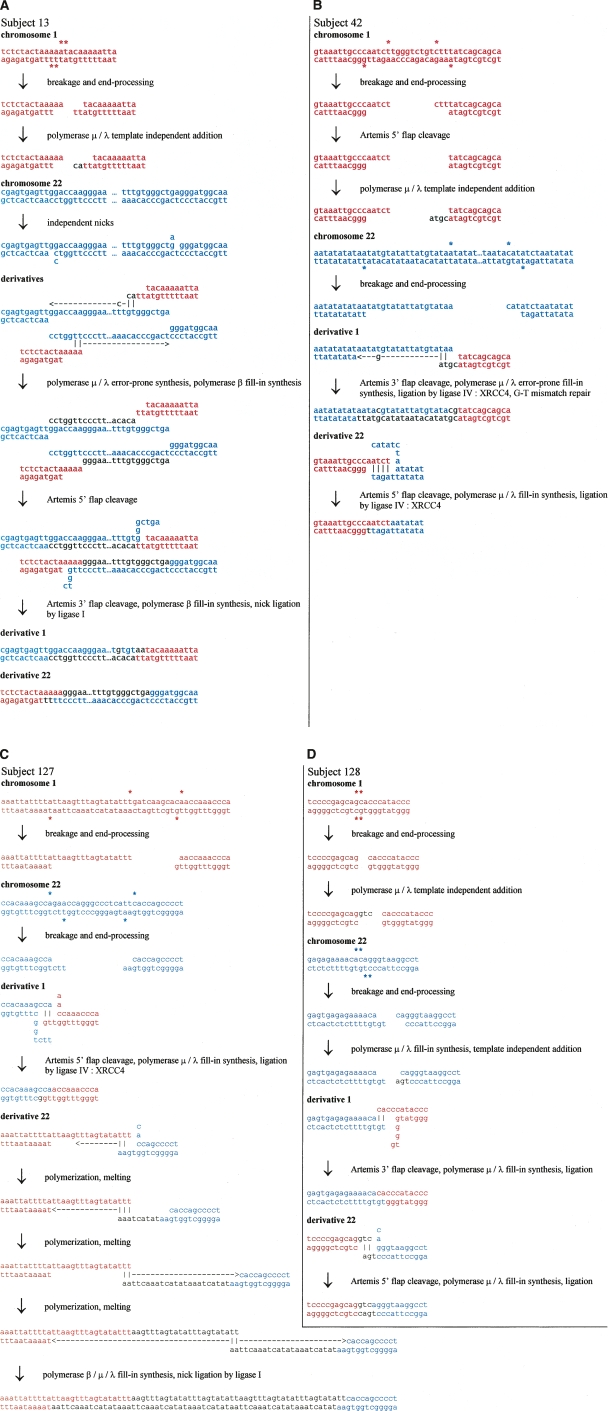
Figure 2.
Possible NHEJ scenarios for four balanced translocations t(1;22)(p36;q13). Sequences presented in red and blue correspond to chromosome 1 and 22 sequence, respectively. (A) Scenario for subject 13. A DSB on chromosome 1 results in a pair of 3′ overhangs—ends with unpaired bases on the 3′ strand. These overhangs invade into separate nicks on chromosome 22 that have opened a little. Polymerase μ or λ polymerizes a few bases on one of the ends, template-independently, and this finds some homology in one of the opened nicks. The polymerase then adds template-dependently, accidentally creating a T–C mismatch in the process. Polymerase β (POLB) fills in the gap. The other 3′ overhang has some microhomology with the other nick on chromosome 22, and polymerase μ or λ polymerizes a few bases template-dependently before ceding to polymerase β. The polymerases continue until they reach the original position of the other nick, and the original chromosome 22 DNA strands are separated along the way. The remaining flaps from the microhomology search are cleaved by ARTEMIS and the nicks sealed by ligase I. The result is a long duplication of chromosome 22 sequence. (B) Scenario for subject 42. A DSB on chromosome 22 results in a 3′ overhang end—an end with unpaired bases on the 3′ strand—and a 5′ overhang end—an end with unpaired bases on the 5′ strand. Chromosome 1 also has a DSB which results in a 3′ overhang end and a 5′ overhang end. The 5′ overhang end is resected by ARTEMIS and extended by polymerase μ or λ template-independently, creating a 3′ overhang end. This end has some microhomology with the 3′ overhang end of chromosome 22, and the microhomology is extended by polymerase μ or λ, which accidently polymerizes a G–T mismatch during the process. The mismatch is aberrantly repaired, resulting in a one-base mutation. The remaining 3′ overhang end of chromosome 1 has some microhomology in the 5′ overhang end of chromosome 22 after some opening. Any remaining flaps are cleaved by ARTEMIS, single-stranded DNA gaps are filled by polymerase μ or λ, and nicks are sealed by ligase IV:XRCC4. (C) Scenario for subject 127. A DSB on chromosome 1 generates two 3′ overhang ends—ends with unpaired bases on the 3′ strand—while a DSB on chromosome 22 results in a 3′ overhang end and a 5′ overhang end. One of the chromosome 1 ends has some microhomology with the 3′ overhang end from chromosome 22 after it has opened a little. The 5′ flaps are resected by ARTEMIS and the single-stranded DNA gaps filled by polymerase μ or λ and sealed by ligase IV:XRCC4. The remaining chromosome 1 3′ overhang end has some microhomology on the remaining chromosome 22 end, which is extended by polymerase μ or λ. The ends undergo several rounds of melting and microhomology-mediated polymerization. Any remaining single-stranded DNA gaps are filled by polymerases μ or λ and β, and the nicks sealed by ligase IV:XRCC4 or ligase I. (D) Scenario for subject 128. A DSB on chromosome 1 results in two blunt ends, one of which is extended template-independently by polymerase μ or λ to result in a 3′ overhang end. Another DSB on chromosome 22 results in two 5′ overhang ends. One of these ends is filled template-dependently by polymerase μ or λ, and then an extra base added template-independently, resulting in a 3′ overhang end. The 5′ overhang end of chromosome 22 has some microhomology with the 5′ overhang of chromosome 1 after both have opened a little. The 5′ flap is cleaved by ARTEMIS, the single-stranded DNA gaps filled by polymerase μ or λ, and the nicks sealed by ligase IV:XRCC4. The remaining chromosome 1 3′ overhang end has some microhomology with the remaining chromosome 22 5′ overhang end after it has opened, and the flap cleavage, gap-filling, and sealing proceed as before.